

PTM Search
PTM Search allows users to search for PTMs residing in specific amino acid sequences or motifs, e.g. [ACM][AS](nt)[AS] - searching a consensus motif of Stromal Processing Peptidase (PMID 26371235, Fig. 3B).
In addition, multiple PTMs can be searched to find potential cross-talk between PTMs, e.g. D(nt)X{1,10}K(ub)
Please consult help section below for detailed information
A downloadable tutorial is available here
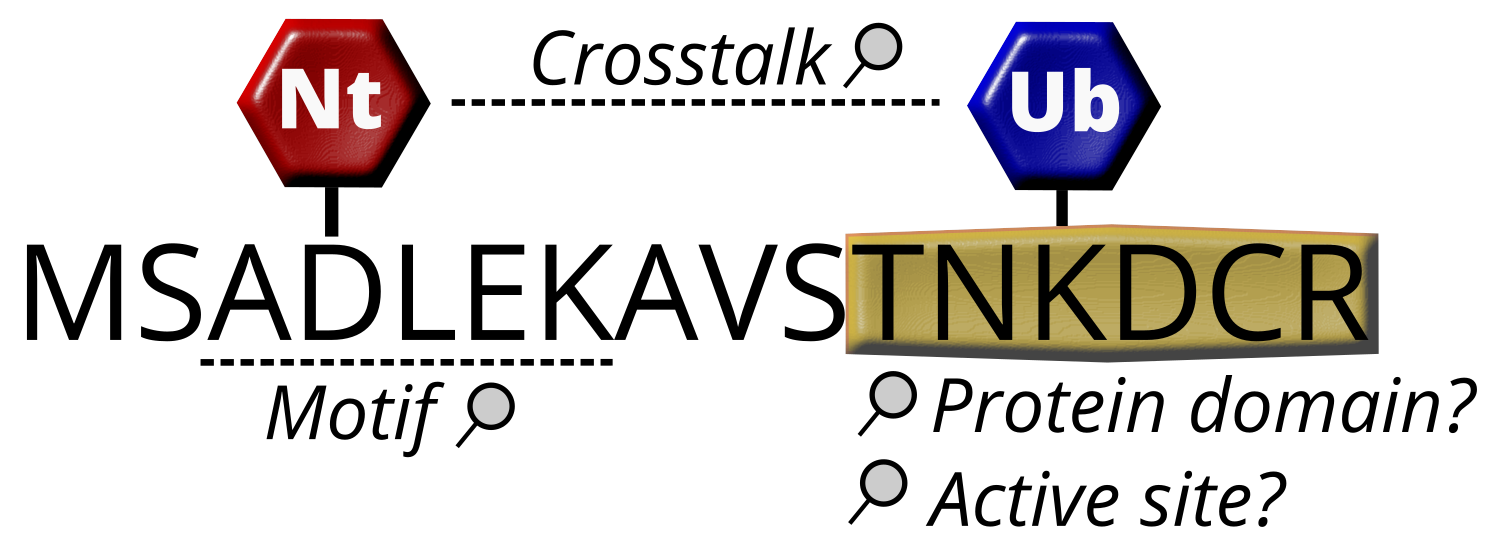

PTMs
Each search must include at least one PTM, denoted by two letters in brackets after the affected amino acid
For example, M(mo) will search for oxidation on Methionine
Available PTM types
| ac | Acetylation |
| acy | S-Acylation |
| ca | Carbonylation |
| cn | S-cyanylation |
| co2 | Carbamylation |
| cr | Crotonylation |
| ct | C-terminus proteolysis |
| epg | Ethanolamine phosphoglycerylation |
| fuc | O-Fucosylation |
| gsh | S-Glutathionylation |
| hib | 2-Hydroxyisobutyrylation |
| hyp | Proline hydroxylation |
| lip | Lipoylation |
| mal | Malonylation |
| mar | MARylation |
| me1 | Monomethylation |
| me2 | Dimethylation |
| me3 | Trimethylation |
| mox | Methionine Oxidation |
| myr | Myristoylation |
| ng | N-glycosylation |
| nt | N-terminus Proteolysis |
| nta | N-terminal Acetylation |
| nub | N-terminal Ubiquitination |
| og | O-GlcNAcylation |
| ox | Reversible Cysteine Oxidation |
| pgk | 3-Phosphoglycerylation |
| ph | Phosphorylation |
| sno | S-nitrosylation |
| so | S-sulfenylation |
| suc | Succinylation |
| sumo | SUMOylation |
| ub | Ubiquitination |
More than one PTM type for the specified amino acid may be searched using the | character for an "or" combination, or the & character for an "and" combination
A search for M(mo|ub) would return results where EITHER oxidation or ubiquitination on the Methionine were found
A search for M(ub&nt) would return results where BOTH ubiquitination and N-terminal PTMs have been found on the Methionine
It is possible to use xx as a wildcard
A search for M(xx) would return any PTM type found on Methionine
Sequence
Search sequences may be a simple string of letters
A search for MK(ub)S would return proteins containing the amino acid sequence MKS for which we have experimental data showing ubiquitination on the Lysine
More complex search strings may be formed using regular expressions
Allowed regex terms are listed below, and should be familiar to those who have dealt with regular expressions before
Regex operators
| [ ] | Match any of the characters contained within the square brackets A search for MA[ER]T will return both MAET and MART (but not MAT) |
| X or . | Match any character A search for MAXT or MA.T will return MAET, MART, etc. |
| ? | Match zero or one occurrence of the character A search for MAE?T will return both MAET and MAT |
| + | Match one or more occurrences of the character A search for MAE+T would return MAET, MAEET, MAEEET, etc, but not MAT |
| * | Match zero or more occurrences of the character A search for MAE*T would return MAT, MAET, MAEET, etc. |
| { } | Specify the number of occurrences of the character A search for MAE{3}T would return MAEEET Two comma-separated numbers can be used to specify a range A search for MAE{2,4}T would return MAEET, MAEEET, and MAEEEET One number and a comma creates an open-ended range A search for MAE{,7} would match between zero and seven E A search for MAE{3,} would match three or more E |
Regex operators which describe the number of repetitions of a character may not be used on the PTM terms in a search string
However, the X/. and [ ] operators may be used for the amino acid of a PTM
A search for MX(nt)D would return proteins containing any sequence M-D for which experimental data exists showing an N-terminal PTM on the unspecified middle amino acid, such as MC(nt)D, ML(nt)D
A search for S[ST](ph)T would return results containing both SS(ph)T, and ST(ph)T
Search Output
Any proteins will be displayed for which experimental data exists matching the search string
The protein amino acid sequence surrounding the search result point will be shown, with all PTMs in the database in this range highlighted, whether or not they were requested in the search term
The size of sequence substring to display can be selected in the Sequence Length search field
All proteins in the table link to their individual protein page which shows the entire sequence and all PTMs
The result table may be sorted - in order to view certain results together, select the checkboxes by the relevant rows and sort on the checkbox column to pull all those selected to the top