

Plant PTM Viewer

Plant PTM Viewer is a centralized resource for plant post-translational modifications (PTMs) intuitive for wet- and dry-lab scientists.
Plant PTM Viewer provides innovative tools to analyze the potential role of PTMs for specific proteins or in a broader systems biology context.
Plant PTM Viewer is an open repository and accepts newly peer-reviewed plant PTM data - for more information click here
Below we provide a brief outline of Plant PTM Viewer features and current PTM data statistics.
Downloadable tutorials can be found here.
Features
Protein Search
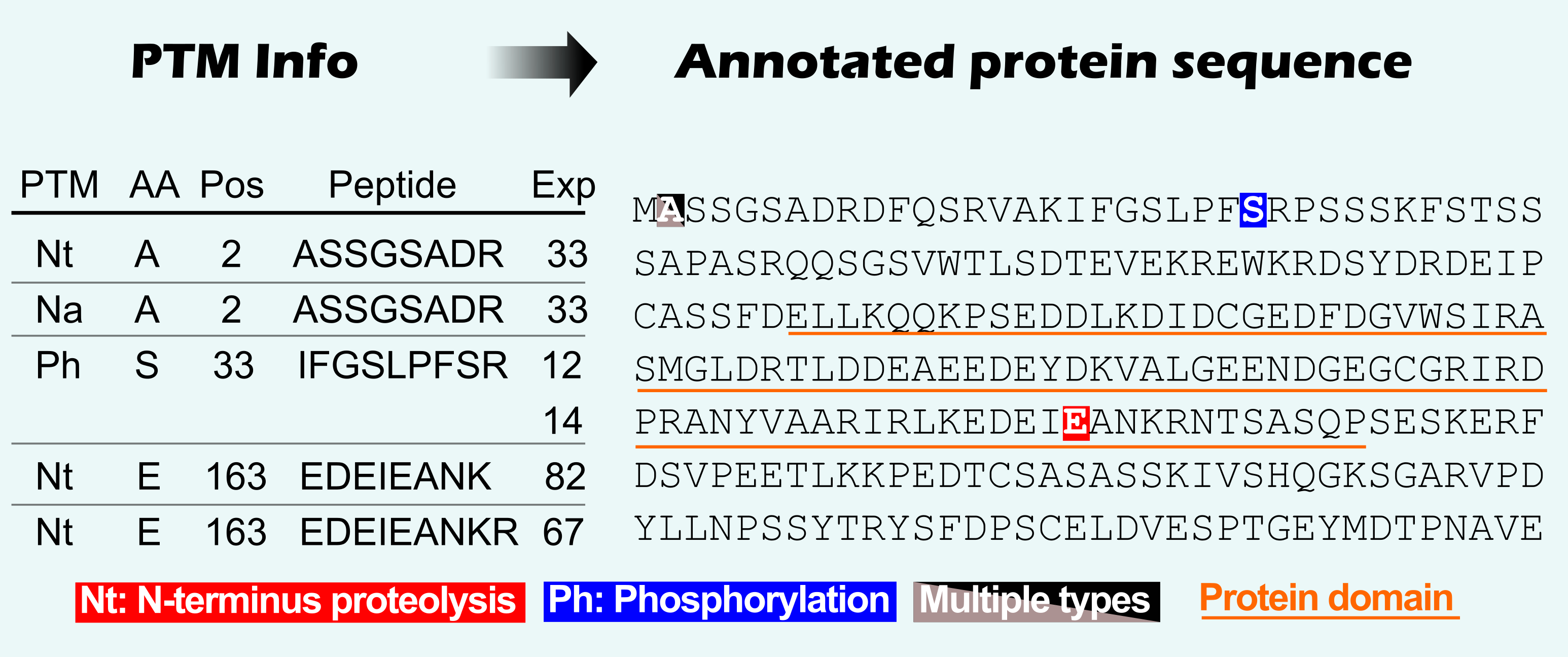

Query your protein of interest via protein identifier, description, or protein sequence and view the complete PTM overview mapped on the protein sequence.
Protein domains and active sites can be shown and compared relative to PTM sites.
Additional PTM information, such as differential statistics and PTM site localisation probabilities are provided if described in the respective publication.
Protein domains and active sites can be shown and compared relative to PTM sites.
Additional PTM information, such as differential statistics and PTM site localisation probabilities are provided if described in the respective publication.
PTM Search
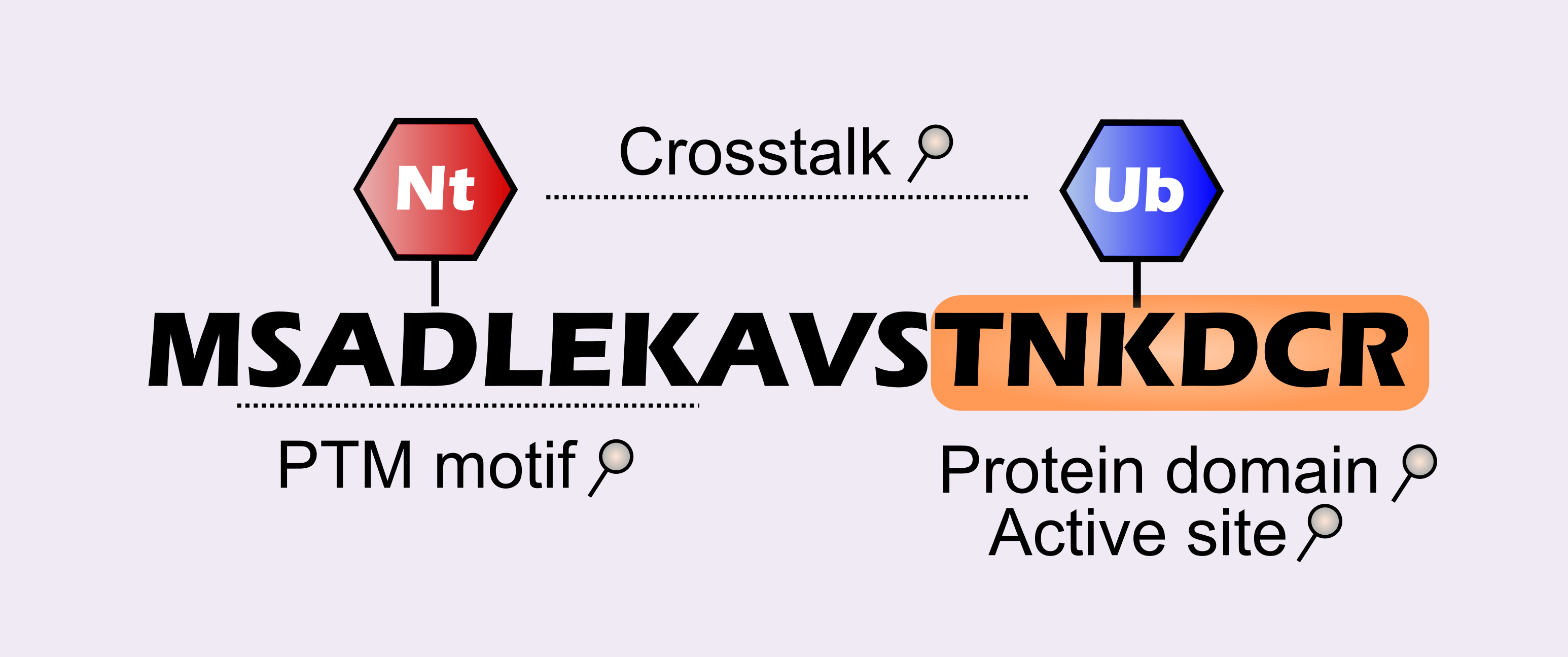

Query your PTM type in a desired regular expression amino acid sequence. Useful to find PTMs residing in specific sequence motifs or discover co-occurrence or interplay between two PTM types.
In addition, PTMs can be searched that map to specific protein domains or active sites.
For instance:
LS(ph)M Phosphorylated Ser after Leu, before Met?
K(ac&ub) Lys both acetylated and ubiquitinated?
T(nt)M(mo) Thr cleavage followed by Met oxidation?
In addition, PTMs can be searched that map to specific protein domains or active sites.
For instance:
LS(ph)M Phosphorylated Ser after Leu, before Met?
K(ac&ub) Lys both acetylated and ubiquitinated?
T(nt)M(mo) Thr cleavage followed by Met oxidation?
PTM BLAST
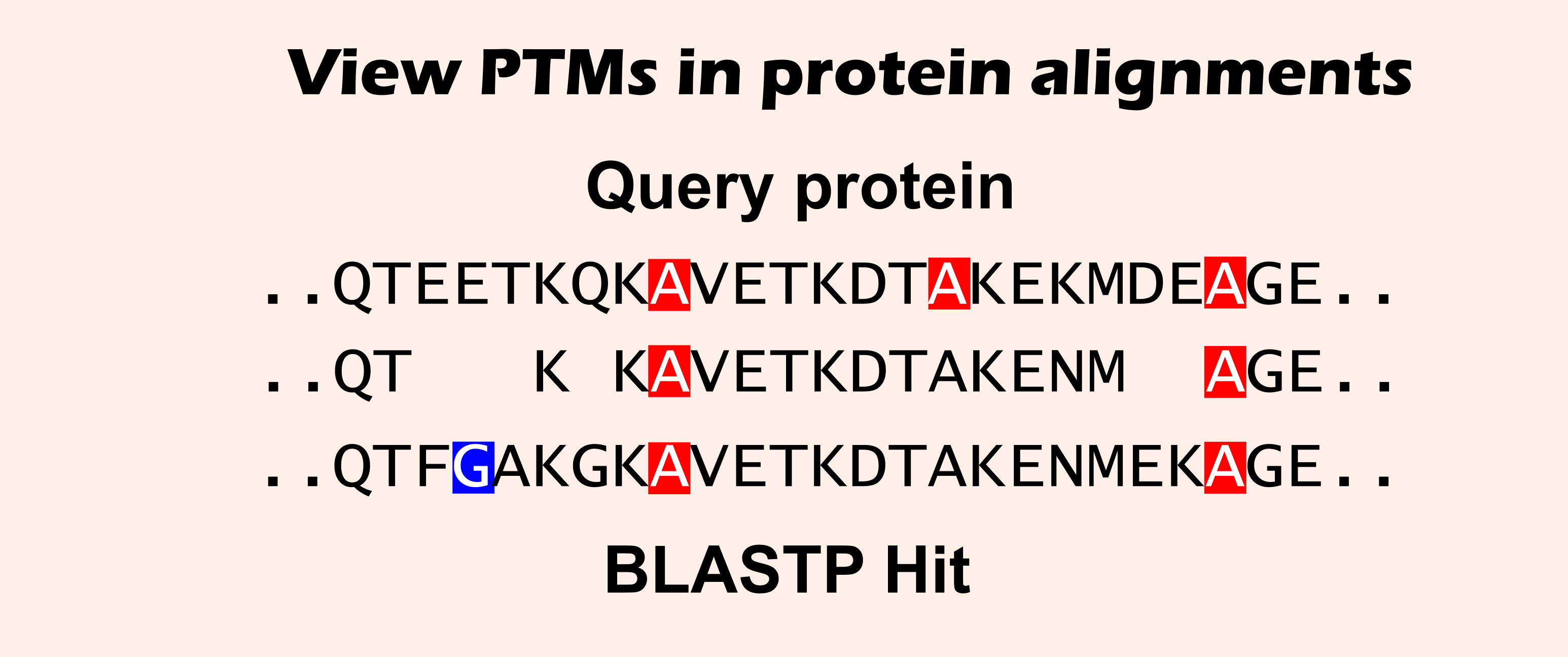

Input a sequence with (or without) specified PTMs and protein BLAST against Plant PTM Viewer data.
In the resulting alignments, PTMs will be displayed and aligned PTMs can be tracked down across species and within protein families. This can reveal PTM sites conserved in protein families within or across species.
PTM BLAST of protein sequences, or part thereof, is an option available on each protein overview.
In the resulting alignments, PTMs will be displayed and aligned PTMs can be tracked down across species and within protein families. This can reveal PTM sites conserved in protein families within or across species.
PTM BLAST of protein sequences, or part thereof, is an option available on each protein overview.
LIST ANALYSIS
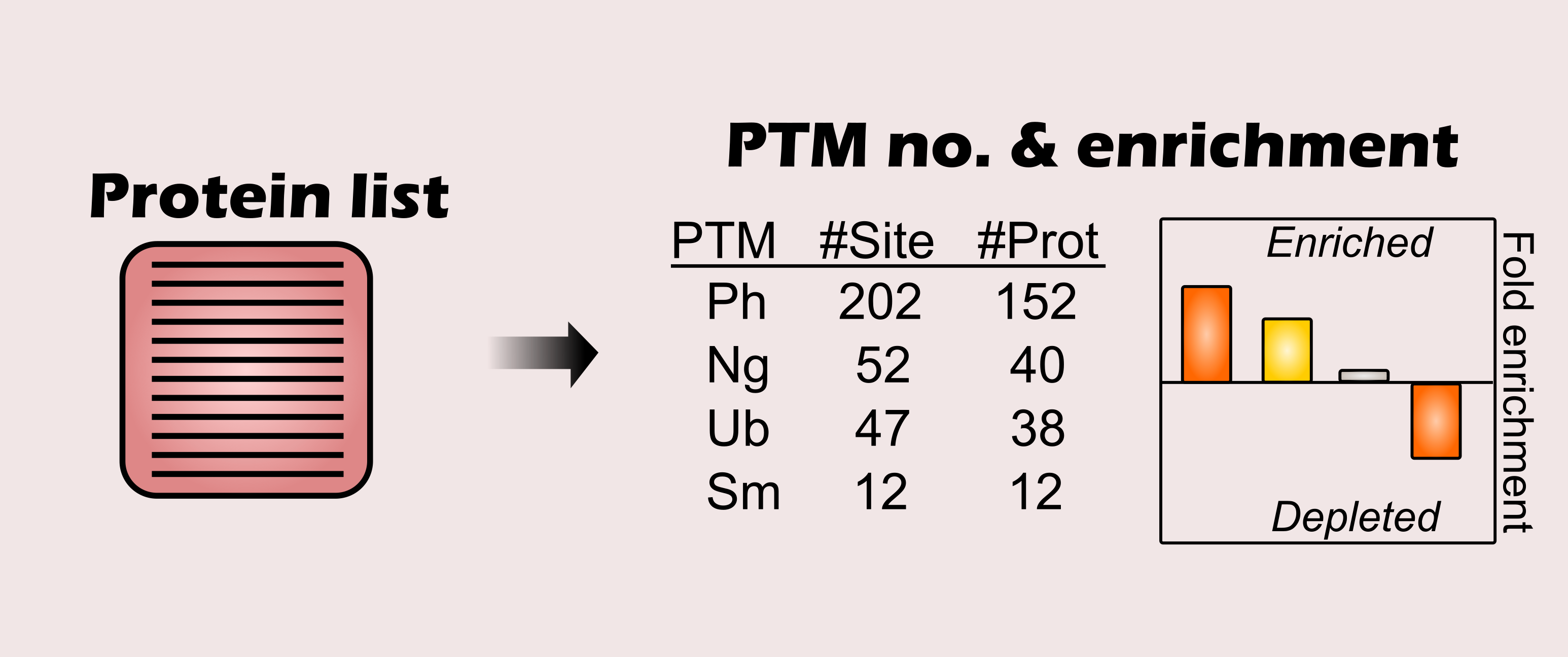

Query a list of proteins for a plant species in order to retrieve the number of modified sites and proteins per PTM. In addition, a PTM set enrichment analysis will be executed for each PTM, thus analyzing the over- or underrepresentation of each PTM in the inputted protein list. All PTM hits and enrichment statistics can be exported - as well as an enrichment graphics.
PTM Viewer Data
Stored PTMs
Loading PTM stats...
Species
Loading species stats...