

PTM Info
Plant PTM Viewer contains data of the 33 PTM types below
Abbreviation
Name
Sites
PTMs
Proteins
Experiments
ac
Acetylation
K
34939
16575
19
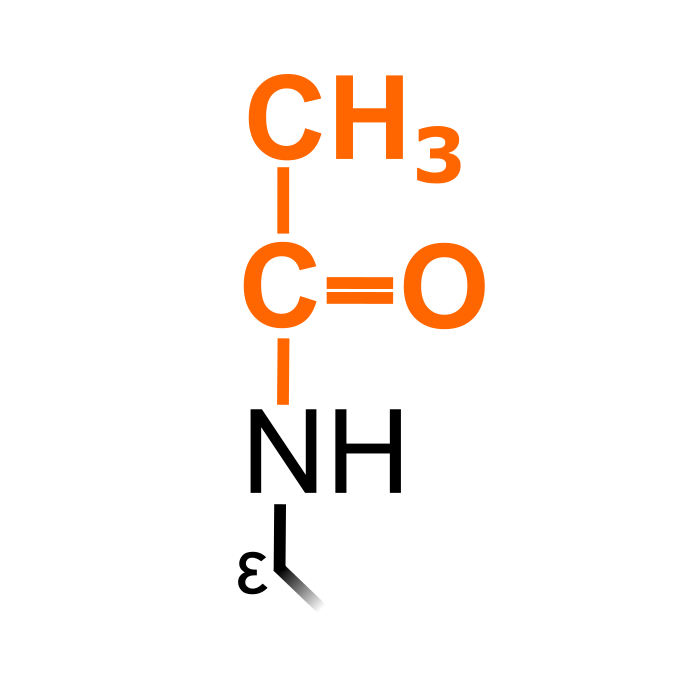
Lysine acetylation is one of the most common PTM in eukaryotes. An acetyl group is transferred from acetyl-CoA to ε-amino groups of Lys, thereby neutralizing the positive charge. Lysine acetylation is reversible and enzymatically catalyzed by Lys acetylases and deacetylases. Note: acetylation on primary amino groups of N-termini are attributed a separate PTM type 'N-terminal acetylation (nta)'.
Δ Mass: +42.011 Da
acy
S-Acylation
C, Rare: N-term
2298
1524
6
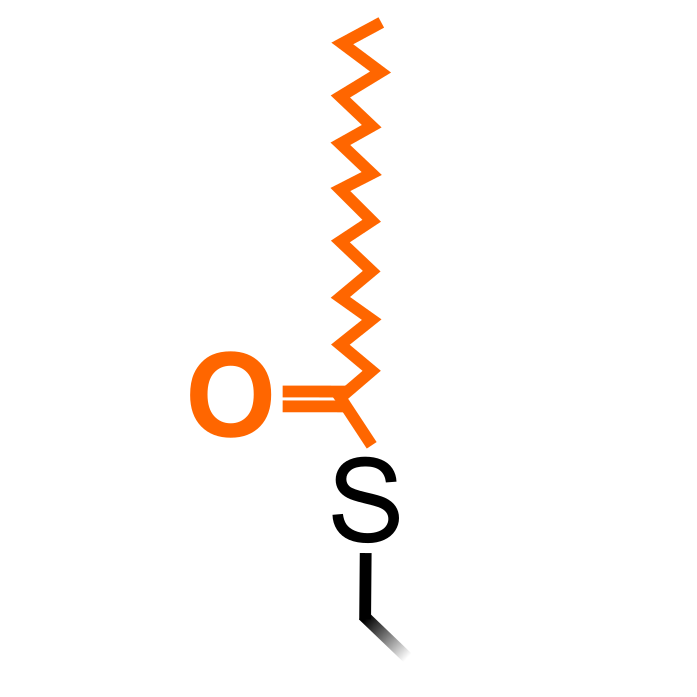
S-acylation is a reversible lipid modification where long-chain lipids are added on Cys thiols via a labile thioester linkage (Chamberlain and Shipston (2015) Physiol Rev 95: 341-376). Most often this long-chain lipid is palmitate, its addition also commonly known as S-palmitoylation, but also others such as stearate (C18:0) and oleate (C18:1). S-acylation increases in protein hydrophobicity that can impact on protein structure, assembly, maturation, trafficking, and function. S-acylation is introduced by S-acyl transferases and removed by S-acyl thioesterases.
Δ Mass: + 238.230 Da (S-palmitoylation)
ca
Carbonylation
R, K, P, T, Y
352
188
3
Irreversible oxidation of protein residues. The addition of carbonyl groups to the proteins may result in protein misfolding, loss of function and degradation by proteasome. Carbonylation products for several amino acids are included in this PTM: Arg (glutamic semialdehyde), Lys (aminoadipic semialdehyde), Pro (5-oxo-proline), Tyr (N-formylkynurenine, kynurenine) and Thr (2-amino-3-ketobutyric acid).
Δ Mass: - 43.053 Da (R), - 1.032 (K), + 13.979 Da (P), - 2.016 Da (T), + 3.995 Da and + 31.990 Da (Y)
cn
S-cyanylation
C
148
132
1
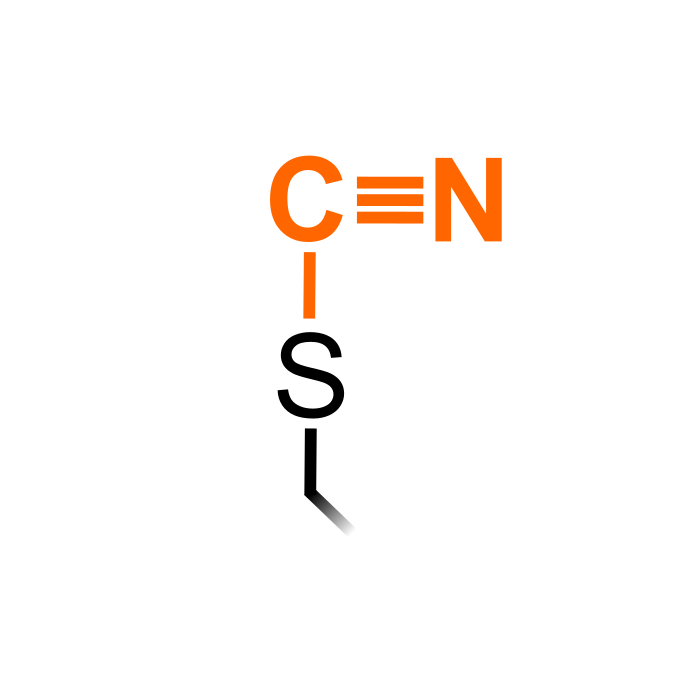
Hydrogen cyanide (HCN) is a gaseous acid that produced as a metabolic byproduct and enzymatically detoxified by the mitochondrial β-CYANOALANINE SYNTHASE (CAS-C1) in plant cells. Reaction of cystine peptides or mixed disulfides with cyanide can form S-cyanylated cysteine. Internal S-cyanylated residues lead to an unstable cyclization form that spontaneously hydrolizes releasing an upstream 2-imino-thiazolidine-4-carboxylyl COOH-terminal peptide. S-cyanylated Cys were identified in a proteomic study in Arabidopsis thaliana (García et al. (2019) Plant Physiol 179: 107-123).
Δ Mass: + 24.995 Da
co2
Carbamylation
K
14
13
1
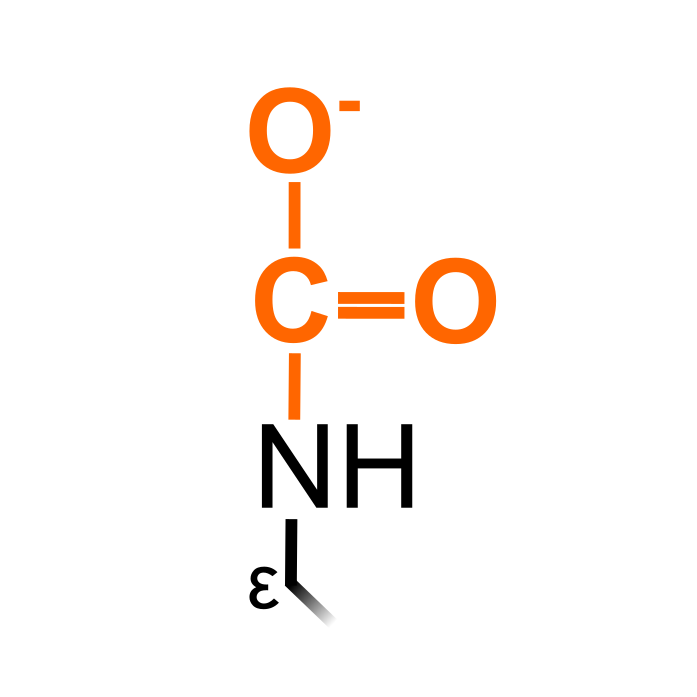
Lys carbamylation is the process of carbon dioxide (CO2) reacting with ε-amino groups of Lys residues, leading to an anionic group that could affect protein function. Carbamate formation has already been suggested as biologically relevant PTM decades ago (George H Lorimer (1983) Trends Biochem Sci 8, 65–68). However, the carbamates are labile and readily reversed, requiring dedicated MS purification and covalent trapping techniques. This was described in Arabidopsis thaliana by Linthwaite et al. (2018, Nat Commun 9: 3902) using triethyloxonium tetrafluoroborate, identifying high confident Lys carbamylation sites in proteins aside from the the well-characterized RuBisCO carbamylation.
Δ Mass: + 42.982 Da
cr
Crotonylation
K
52183
15404
3
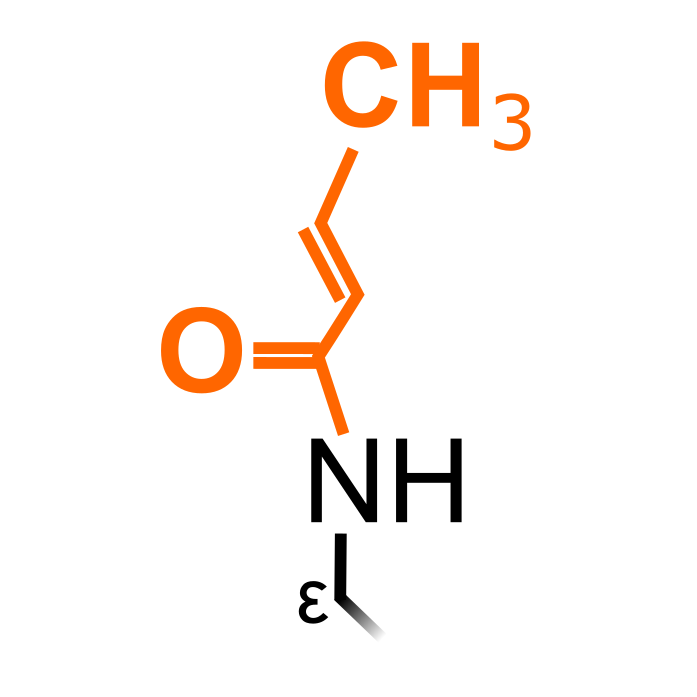
Lysine crotonylation has been discovered in various eukaryotes and is best-known to modify histone proteins. Crotonylation can neutralize the positive charge of the ε-amino group of lysine and has been associated with an active state of gene promoters. In more recent years, crotonylation has been described to also target non-histone proteins.
Δ Mass: + 68.026 Da
ct
C-terminus proteolysis
Any
2483
1803
1
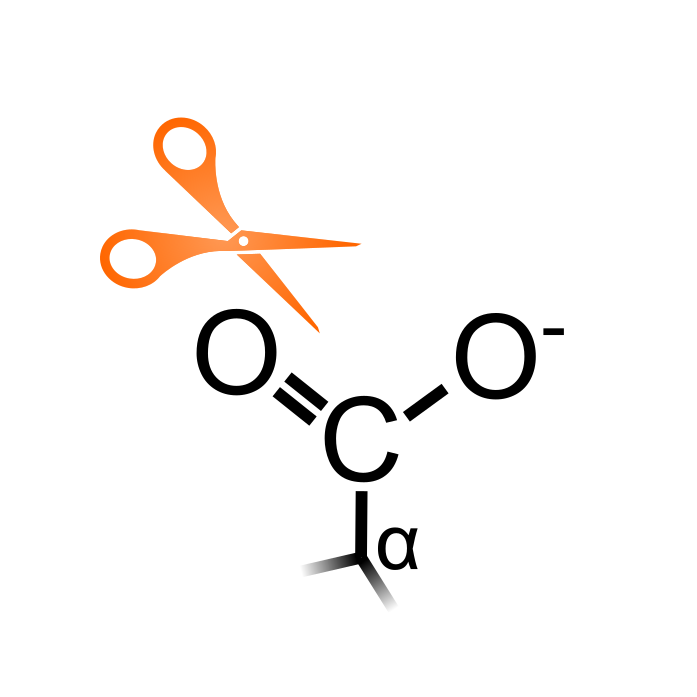
Novel C-termini can be exposed within proteins by proteases that cleave peptide bonds. Proteomic techniques have been developed to enrich in vivo C-terminal peptides, indicative of neo-C-termini generated by cleavage events. Profiling of C-terminal peptides has far lagged behind compared to N-terminal peptide profiling due to the low reactivity of C-terminal carboxyl groups and lower ionization of C-terminal peptides. However, mass spectrometry studies are starting to profile C-termini in various species. For instance, a method termed BaSCX with LysargiNase digestion identified approximately 1,000 of C-termini in tomato (Li et al. (2020) Anal Chem 92:4742-4748). Note that the C-terminus of biochemically verified hormone peptides are also considered here in this PTM type.
Δ Mass: -
epg
Ethanolamine phosphoglycerylation
E
9
9
1
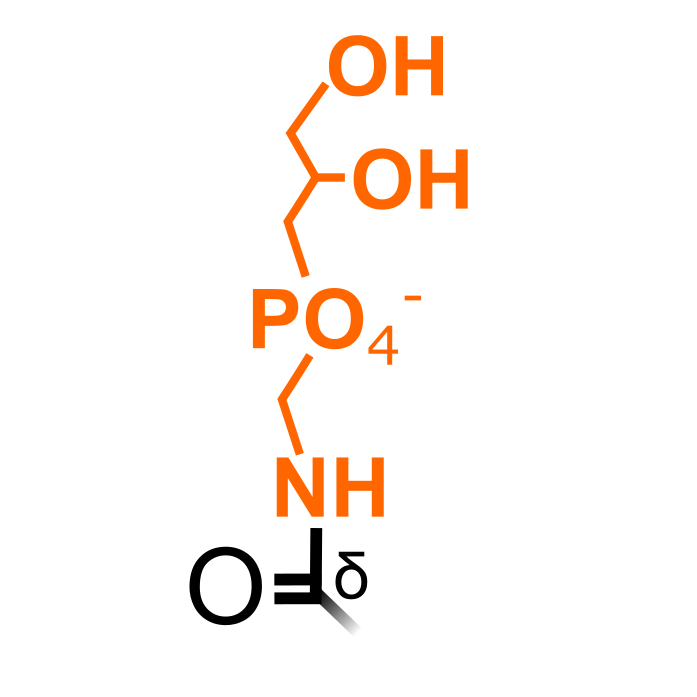
Initially, two Glu resides of Elongation factor 1 alpha (EF-1α) were found to be posttranslationally modified by ethanolamine-phosphoglycerol (epg) groups (Whiteheart et al. (1989) J Biol Chem 264:14334-14341). Also in Arabidopsis thaliana, incorporation of isotopic-labeled ethanolamine was observed in EF-1α, suggested to facilitate its subcellular distribution (Ransom et al. (1998) Plant Physiol 117:949-960). In the Plant PTM Viewer 2.0 update paper, we identified epg groups (and ethanolamine) on Glu301 on EF-1α using open searches.
Δ Mass: + 197.045 Da
fuc
O-Fucosylation
S,T
718
420
1
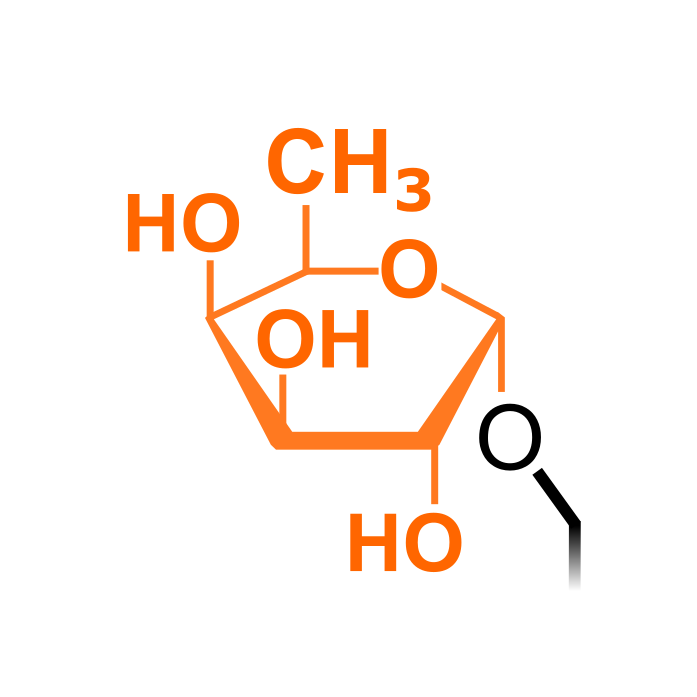
Fucose can be attached via O-linkage to the hydroxyl groups of serine or threonine. In Arabidopsis thaliana, SPINDLY (SPY) exerts a O-fucosyltransferase activity and was shown to target DELLA proteins (Zentella et al., 2017). A large-scale enrichment study using a lectin pull-down identified hundreds of O-fucosylated sites in Arabidopsis.
Δ Mass: + 146.080 Da
gsh
S-Glutathionylation
C
66
46
1
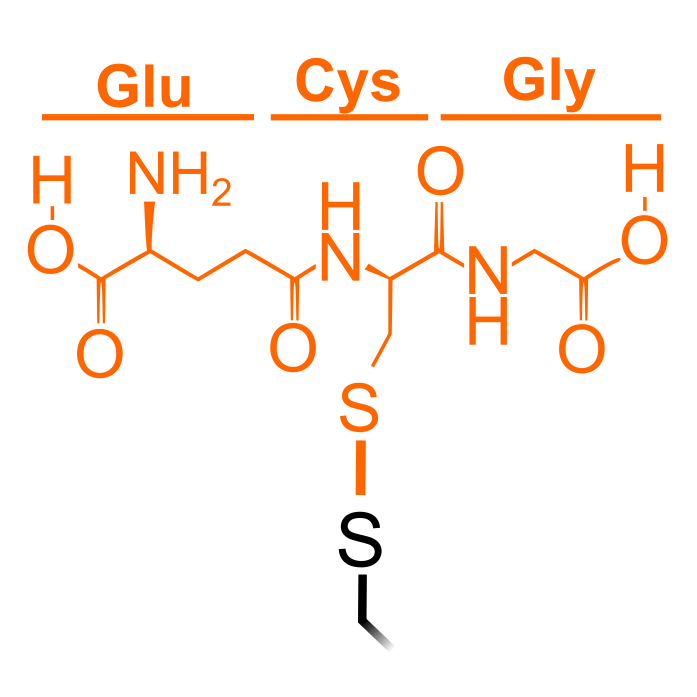
S-Glutathionylation is the reversible disulfide linkage of the low-molecular-mass thiol glutathione (GSH) to Cys thiols. It is transferred by glutathione S-transferases (GSTs) and reduced by glutaredoxins (Grxs). S-Glutathionylation can have a protective role by preventing overoxidation or proteolysis. S-glutathionylation is reduced by DTT/TCEP typically used during proteomic protocols and thus requires dedicated, non-reducing workflows for detection.
Δ Mass: +305.068 Da
hib
2-Hydroxyisobuturylation
K
133998
34884
7
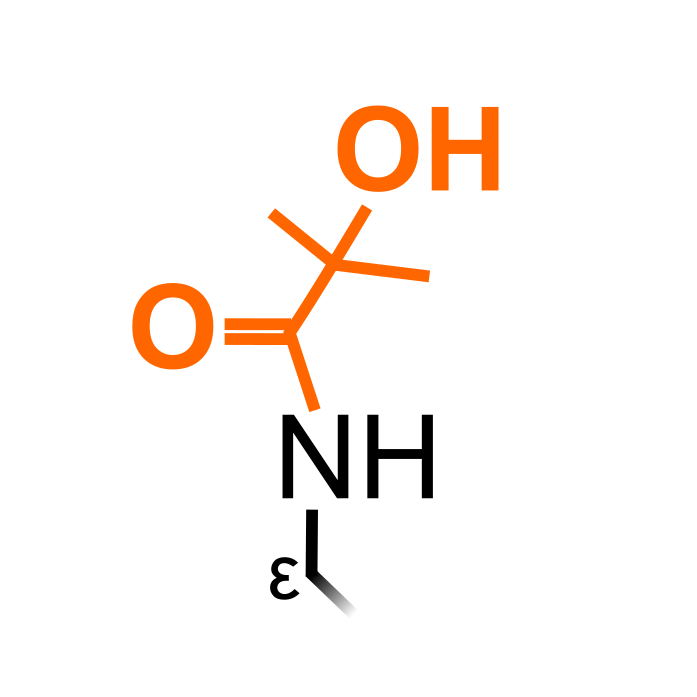
Lysine 2-Hydroxyisobutyrylation (Khib) was identified as a novel histone mark, introduced by 2-hydroxyisobutyryl-CoA (Dai et al., 2014, Nat Chem Biol 10:365-370) . The short-chain acylation of Lys side-chains neutralizes the positive charge of Lys, but is also larger in size than acetyl and contains a hydroxyl group that could form hydrogen bonds with other molecules. Next to histones, several non-histone proteins are contain Khib sites - and several MS studies have been performed in plants.
Δ Mass: +86.03 Da
hyp
Proline hydroxylation
P
133
82
1
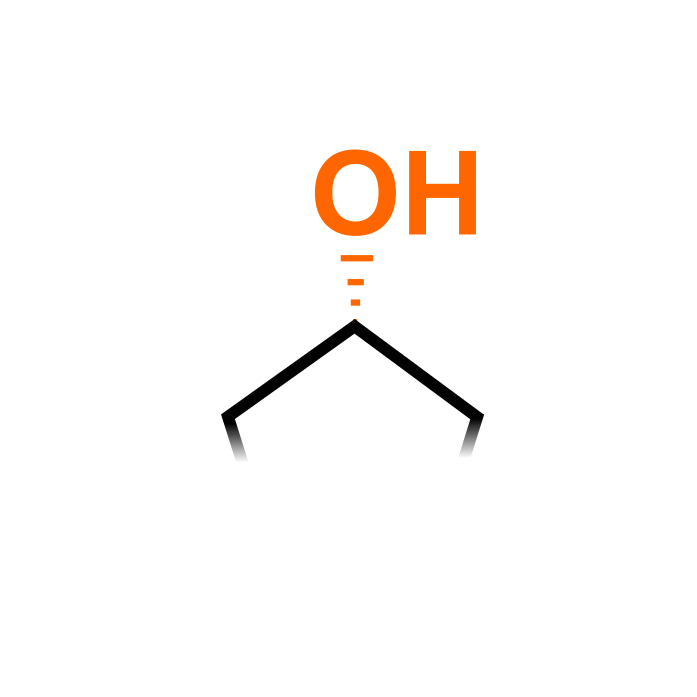
Hydroxyproline (Hyp) is generated by oxidation of Pro residues and is catalyzed by prolyl-4-hydroxylases in plants. Proline hydroxylation follows a certain preference or code, e.g. they are more likely to be hydroxylated when following Ala, Gln, Hyp, Pro, Ser, Thr and Val residues. Hydroxyproline frequently occurs on cell wall proteins, secreted peptide hormones and especially hydroxyproline-rich glycoproteins, where Hyp are subjected to O-glycosylation (e.g. in PTM search 'P(hyp&og)'.
Δ Mass: + 15.995 Da
lip
Lipoylation
K
2
2
1
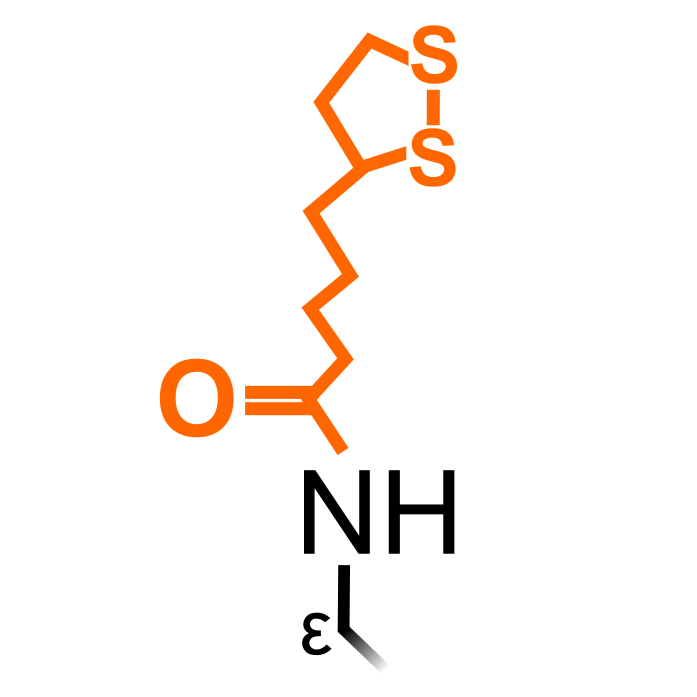
Lipoylation is a rare Lys PTM and has been observed on a handful of core metabolic enzymes. Due the large size of the lipoyl moiety, it can impact protein structure and provide a "swinging arm" function for enzymatic reactions (Rowland et al. (2018) Curr Opin Chem Biol 42:76-85). In the Plant PTM Viewer 2.0 update paper, we identified lipoylation on plant glycine cleavage H-protein Lys in Arabidopsis. This lipoylation acts an essential shuttle in the enzymatic catalysis of the Glycine Cleavage System, and a first structure of lipoyl-Lys was observed in a plant GSH H-protein from pea (Pares et al. (1994) PNAS 91:4850-4853).
Δ Mass: + 188.033 Da
mal
Malonylation
K
11710
5599
3
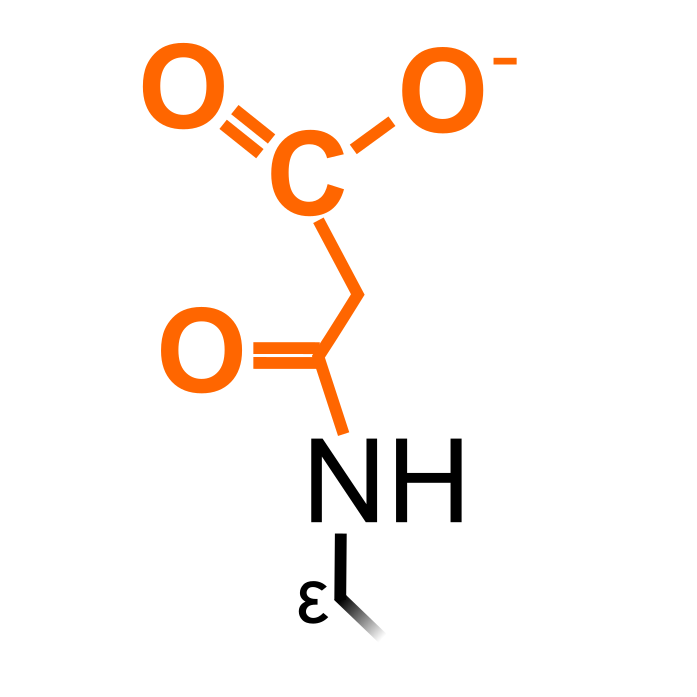
Lysine malonylation (Kmal) is derived from malonyl-CoA and was first discovered in a Lys succinyl dataset and confirmed MS in HeLa cells and E. coli (Peng et al. (2011) Mol Cell Proteomics 10: M111.012658). MS studies enriching malonylated peptides have been performed in plants and indicated high overlap with acetylation and succinylation sites. In addition, the positively charged residues Lys and Arg frequently flanked Kmal.
Δ Mass: +86.000 Da
mar
MARylation
S,E,D,R,K,Y,H,C
44
6
1
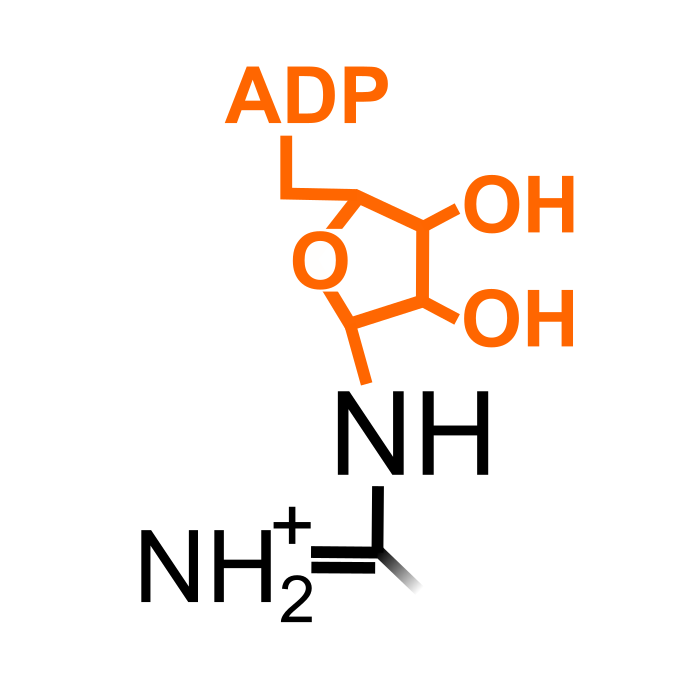
Protein ADP-ribosylation is the reversible covalent attachment of ADP-ribose from NAD+ on acceptor proteins. Addition of a single ADP-ribose unit is referred to as mono(ADP-ribosyl)ation or MARylation. MARylation can target a wide range of amino acids. Plant protein MARylation was described before by bacterial effectors (for instance Fu et al. (2007) Nature 447: 284-288), but recently two zinc finger transcription factors were shown to be MARylated by a non-canonical ADP-ribosyltransferase SRO2 in Arabidopsis (Kong et al. (2021) Mol Cell 81: 4591-4604).
Δ Mass: + 541.061 Da
me1
Monomethylation
R, K, rare: C, N-terminus and C-terminus
961
657
3
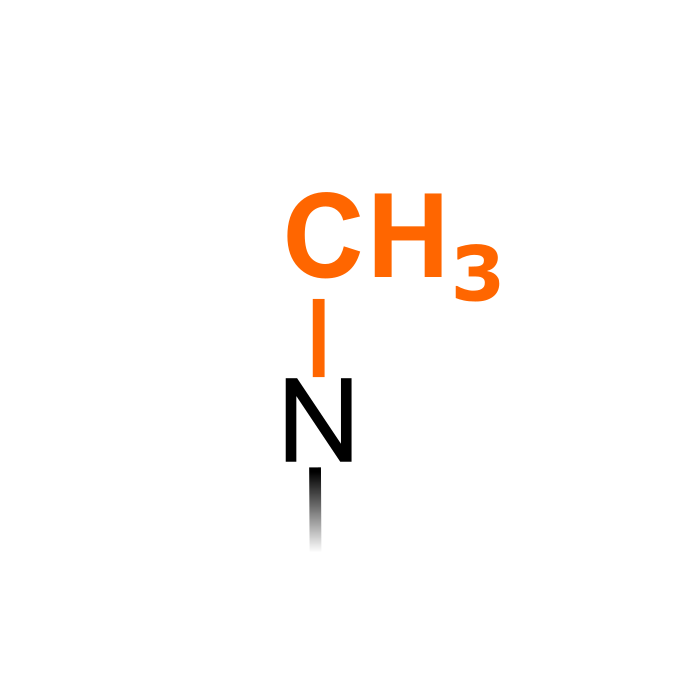
Methylation is catalyzed by lysine and arginine methyltransferases that add a methyl moiety Lys and Arg respectively. S-adenosyl methionine (SAM) is the primary methyl group donor. Methylation increases the hydrophobicity of the protein. Methylation is known to be involved in epigenetic regulation by targeting histones. In few cases methylation of non-Arg/Lys residues has been described, for instance S-methylcysteine has been observed in Chlamydomonas Rubisco (Taylor et al. (2001) J Biol Chem, 276:48159-48164). Note that N-terminal monomethylation is considered here a separate PTM type.
Δ Mass: +14.016 Da
me2
Dimethylation
R, K
803
535
2
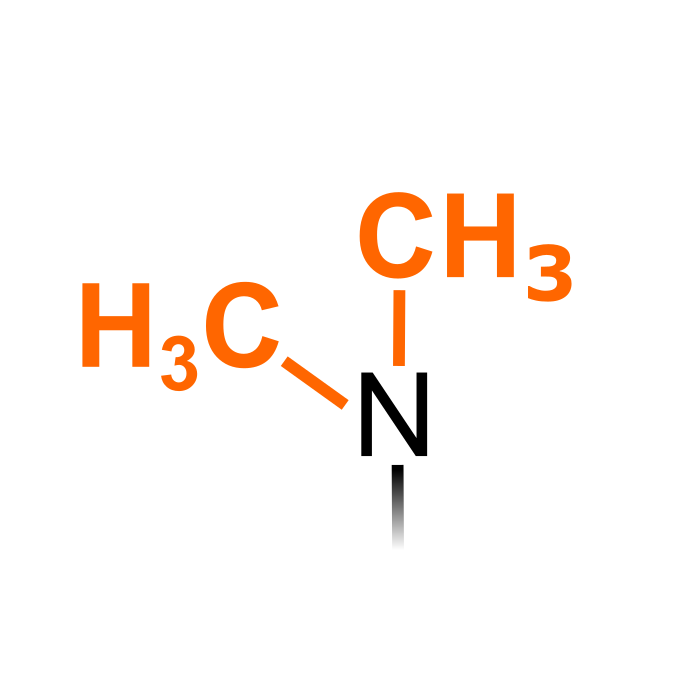
Lysine dimethyl (Kme2) is catalyzed by protein lysine methyltransferases (PKMTs), while three types of protein arginine methyltransferases (PRMTs) can yield asymmetric dimethyl arginine (aDMA) or symmetric dimethyl arginine (sDMA) residues. Dimethylated residues can be targeted via immunopurification strategies. Note that N-terminal dimethylation is considered here a separate PTM type.
Δ Mass: +28.031 Da
me3
Trimethylation
K
181
107
1
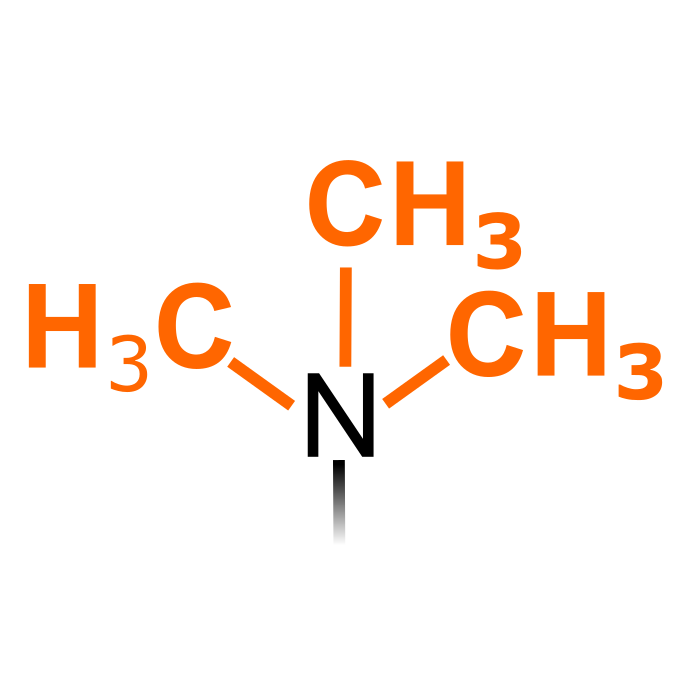
Lysine trimethyl (Kme3) is catalyzed by protein lysine methyltransferases (PKMTs). Trimethylated residues can be targeted via immunopurification strategies. Caution as the the monoisotopic mass differs only 0.036 Da with acetylation that can occur on Lys residues. Note that N-terminal trimethylation is considered here a separate PTM type.
Δ Mass: +42.047 Da
mox
Methionine Oxidation
M
1285
920
2
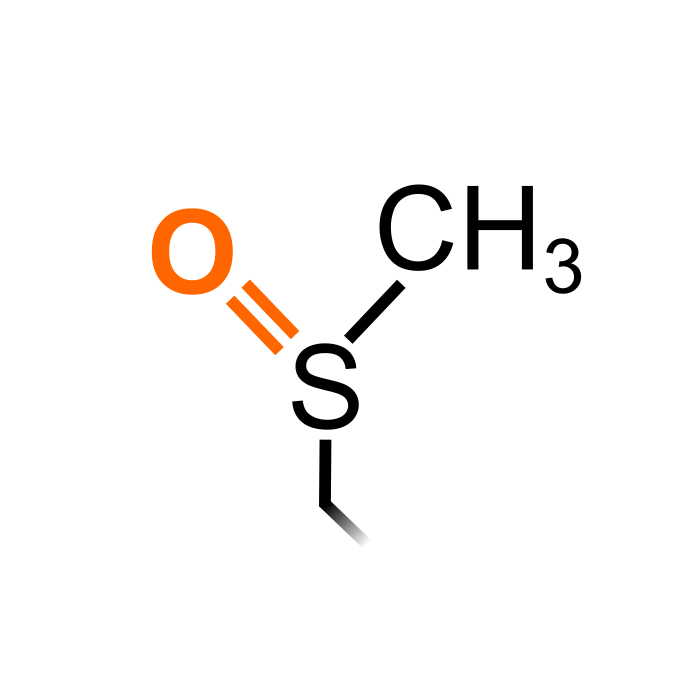
Oxidation of Methionine leads to the formation of methionine sulfoxide that can be reverted by Methionine sulfoxide reductases (MSRs). MSRs on their turn are typically regenerated by thioredoxin or glutaredoxin reducing systems. While the functional knowledge of Met oxidation is in its infancy in plants, its been shown to influence protein activity in few cases. For instance, Met oxidation of the tomato transcription factor NON-RIPENING (NOR) decreases its DNA-binding activity - which is countered by MSRs together defining a Met redox regulation switch influencing fruit ripening (Jiang et al. (2020), Plant Physiol 183: 671-685).
Δ Mass: +15.995 Da
myr
Myristoylation
N-term G
166
166
2
Myristoylation is a lipid N-terminal modification, attaching a myristoyl group to the amino group of N-terminal glycine residues. The reaction is catalyzed by N-myristoyltransferases. It has an important role in membrane targeting and catalyzing protein-protein interactions.
Δ Mass: + 210.198 Da
ng
N-glycosylation
N-!P-[ST], rare: R
5784
3258
10
N-linked (N-)glycosylation refers to the attachment of glycans (oligosaccharides) to the amide nitrogen of Asn. Chemical structures of N-glycans are diverse and vary between species. For instance, late N-glycan maturation steps in the Golgi differ significantly in plants giving rise to complex N-glycans. N-glycosylation is important during protein folding, stability and protein-protein interactions. Some rare cases of Arg glycosylation have been reported in plants (Konishi et al. (2010) Carbohydr Res 345: 787-791; Singh et al. (1995) FEBS Lett 376: 61-64).
Δ Mass: Variable, dependent on N-glycan. Example: + 203.079 Da (HexNAc)
nt
N-terminus Proteolysis
Any
18419
7668
19
Novel N-termini can be exposed within proteins by proteases that cleave peptide bonds. Proteomic techniques have been developed to enrich in vivo N-terminal peptides, indicative of neo-N-termini generated by cleavage events. Plants encode hundreds of proteases divided in distinct classes according their catalytic mechanism. Some proteolytic events are ubiquitous in cells such as co-translational cleavage of the initiator Met (iMet) by aminopeptidases or the cleavage of targeting peptides of proteins targeted to the chloroplast, mitochondria or secretory pathway. Proteolytic fragments can acquire specialized functions (i.e. hormone peptides) or be further degraded. Novel exposed N-termini can be modified by other PTMs or be a hallmark for degradation. Note: N-terminal peptides matching the native protein N-terminus (i.e. iMet) are not considered a PTM.
Δ Mass: -
nta
N-terminal Acetylation
N-terminus
7248
6467
15
N-terminal acetylation (NTA) is a very frequent modification that can take place co-translationally and target the α- amino group of the protein N-terminus. N-terminal acetylation is catalyzed by N-acetyltransferases (Nat) with different specifities. NatA can target Ser, Ala, Thr, Gly and Cys N-terminal residues exposed after N-terminal methionine excision. NatB and NatC can acetylate the initiator Met followed by acidic or hydrophobic residues respectively. In plants, NTA can also occur post-translationally in chloroplasts on the neo-N-terminus generated after transit peptide cleavage. Deacetylases of N-terminal acetylation have not yet been identified and is thus considered irreversible.
Δ Mass: +42.011 Da
nub
N-terminal ubiquitination
N-terminus
24
24
1
While ubiquitination is predominantly identified on Lys sidechains, rare cases of N-terminal ubiquitination have been reported. In the study of Walton et al. (2016, Plant Cell 28: 6-16), 7 N-terminal ubiquitination sites are reported, although this number might increase in the future with improved ubiquitination detection techniques. For instance, the recent UbiSite technology identified a significant portion of N-terminal ubiquitination in human cell lines (Akimov et al. (2018) Nat Struct Mol Biol 25: 631-640). N-terminal ubiquitination has been show to mark proteins for degradation (Coulombe et al. (2004) Mol Cell Biol 24: 6140–6150), and N-terminal acetylation has been suggested to protect from N-terminal ubiquitination and thus degradation.
Δ Mass: +114.04 Da (GlyGly tryptic remnant)
og
O-GlcNAcylation
S, T, Hyp
838
430
1
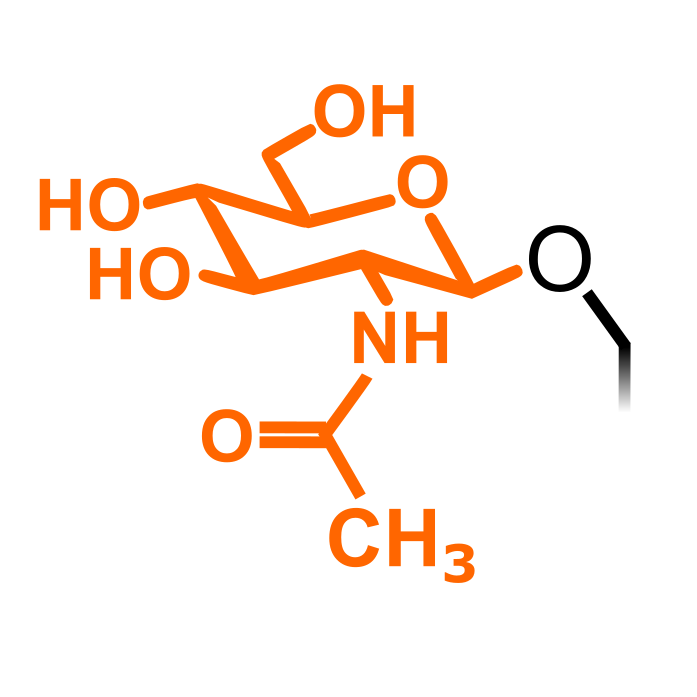
O-GlcNAcylation (O-linked N-acetylglucosamine) can occur on Thr, Ser and hydroxyproline (Hyp), sharing similarities and having interplay with phosphorylation. O-GlcNAcylation often takes place on nucleocytoplasmic proteins, without further attachment of additional sugar moieties. It is induced by O-GlcNAc transferases (OGTs) and removed by O-GlcNAcases (OGAs).
Δ Mass: +203.079 Da (HexNAc)
ox
Reversible Cysteine Oxidation
C
5525
3826
10
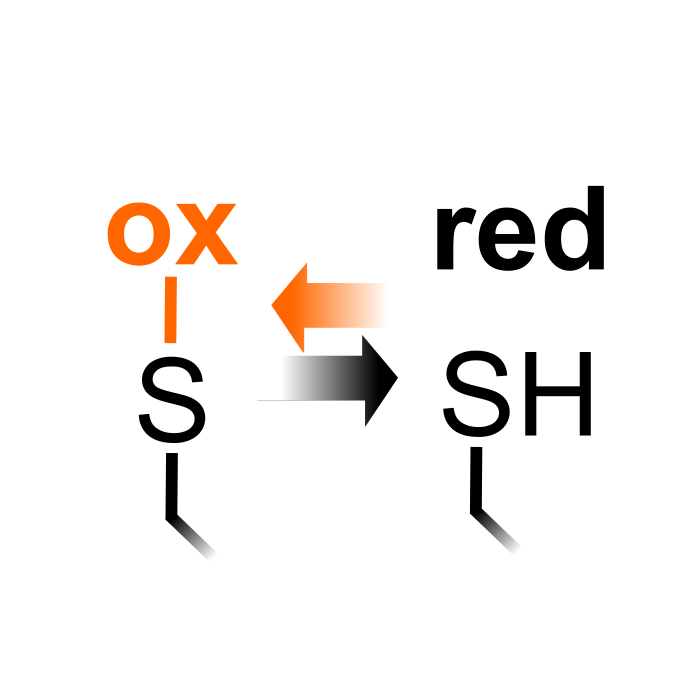
Reversible Cys oxidation is used here as a PTM type to describe multiple reversible Cys oxidation forms (i.e. disulfides, S-glutathionylation, sulfenic acid) that are quantified/studied by reductome studies. In brief, these studies first irreversibly block all free, non-oxidized Cys thiols, after which oxidized thiols are reduced and identified/quantified by mass spectrometry (Willems et al. (2021) Plant Phys 186: 110-124). This can serve as elegant way to quantify the reversible Cys oxidation, the drawback of this indirect detection method is that the precise oxidized form(s) remain elusive and in vitro oxidation has to be avoided at all cost.
Δ Mass: -
pgk
3-Phosphoglycerylation
K
137
93
1
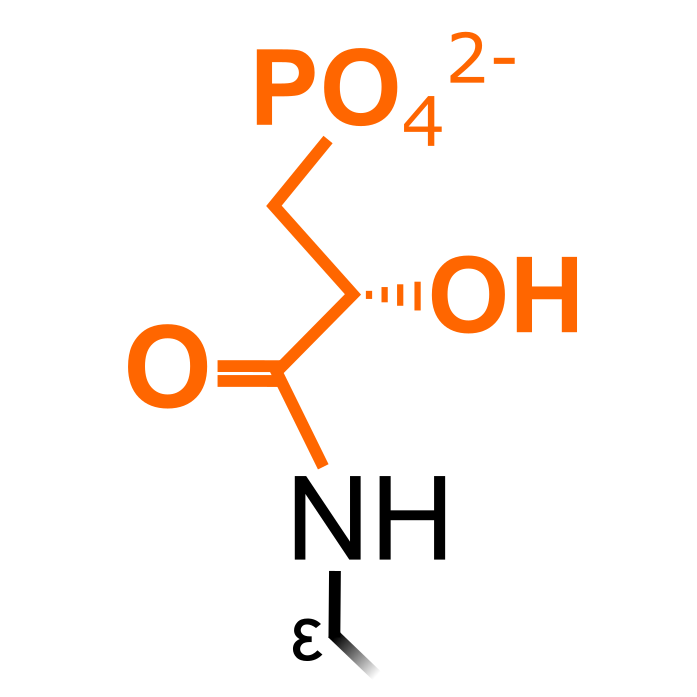
The glycolytic intermediate 1,3-biphosphoglycerate can react with Lys to form 3-phosphoglyceryl Lys (pgK) modifications. This was originally described to largely target and affect the activity of glycolytic enzymes (e.g. GAPDH) in mammals (Moellering and Cravatt (2013), Science 341:549-553). In the Plant PTM Viewer 2.0 update paper, we discovered that plant glycolytic enzymes (and other proteins) are similarly targeted by pgK using open searches, such as GAPDH isoforms in the cytosol and chloroplast.
Δ Mass: + 167.982 Da
ph
Phosphorylation
S, T, Y, Rare: D, N-term
266086
82207
92
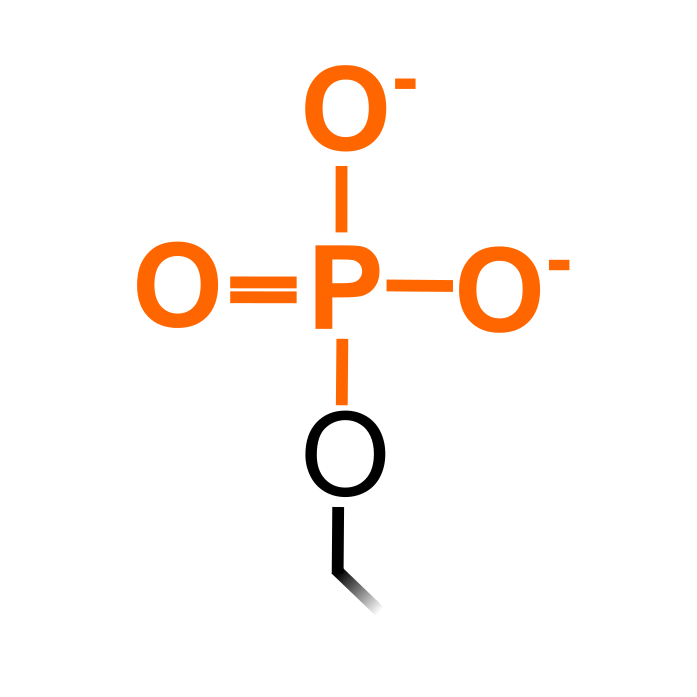
Phosphorylation is by far the best characterized PTM and multiple proteomic studies have profiled phosphorylation in plants. Phosphopeptides are typically enriched by reaction of the negatively charged phosphate groups to metal ions or metal oxide, or using antibodies for enrichment (e.g. phosphotyrosine antibodies). Phosphorylation is mediated by kinases and removed by phosphatases. Many effects on protein function have biochemically characterized for phosphorylation and they can define phosphorylation cascades. Next to Ser, Thr and Typ as major sites, phosphorylation can also take place on other residues such as Asp (Hass et al. (2004) EMBO J 23: 3290–3302). Note that (rare) cases of N-terminal phosphorylation are considered here a separate PTM type.
Δ Mass: +79.966 Da
sno
S-Nitrosylation
C
9026
5924
9
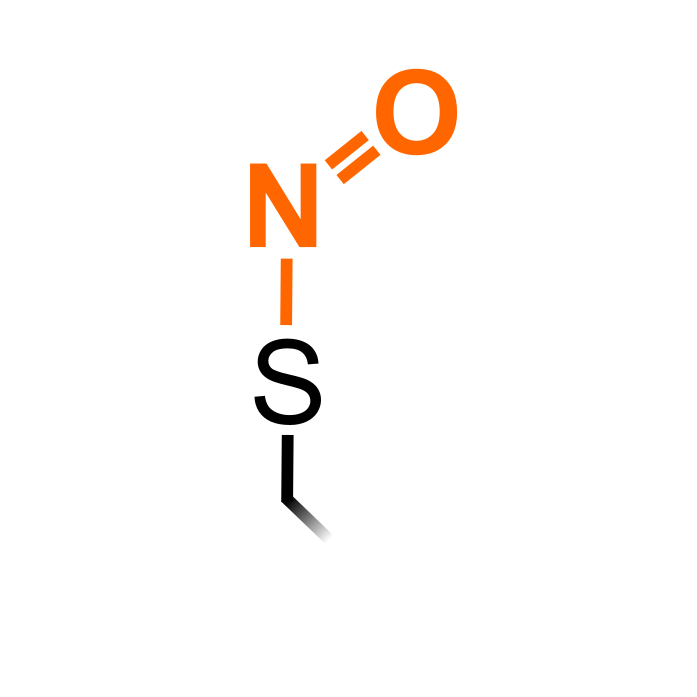
S-nitrosylation (SNO) is a reversible attachment of nitric oxide (NO) to the thiol group of Cys, which is stimulated under NO-inducing conditions. Multiple mechanisms are proposed for SNO formation, including an oxidative pathway with higher oxidized NO forms, radical-mediation formation and metal-catalyzed formation. In addition, trans-nitrosylation can occur, i.e. transferring the nitrosyl moiety from a S-nitrosylated thiol to an acceptor thiol. Denitrosylation is mediated by enzymes such as S-nitrosoglutathione reductase (GSNOR) and thioredoxins. S-Nitrosylation is known to affect protein functions in various manners.
Δ Mass: +28.990 Da
so
S-sulfenylation
C
6489
3871
2
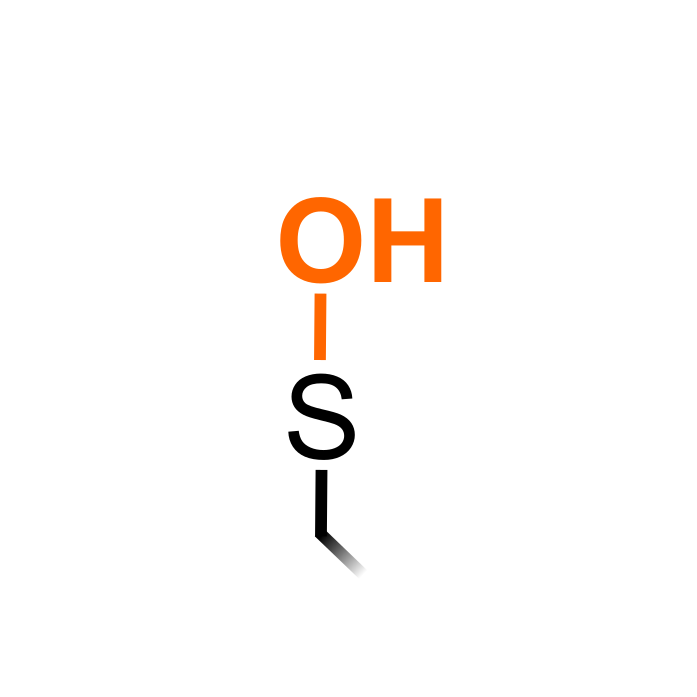
Hydrogen peroxide (H2O2) can oxidize specific protein cysteine thiols to sulfenic acid (SOH), a process known as S-sulfenylation. S-sulfenylation functions as an intermediate on the path toward other redox modifications, such as disulfide formation, S-glutathionylation, and overoxidation to sulfinic (SO2H) and sulfonic (SO3H) acid. With cysteines as reactive residues often participating in active sites of proteins, these modifications can have a direct impact on protein function. Genetic- or chemical-based approach have been developed to trap and identify S-sulfenylated cysteine
s by mass spectrometry.
Δ Mass: +15.995 Da
suc
Succinylation
K
5340
2450
4
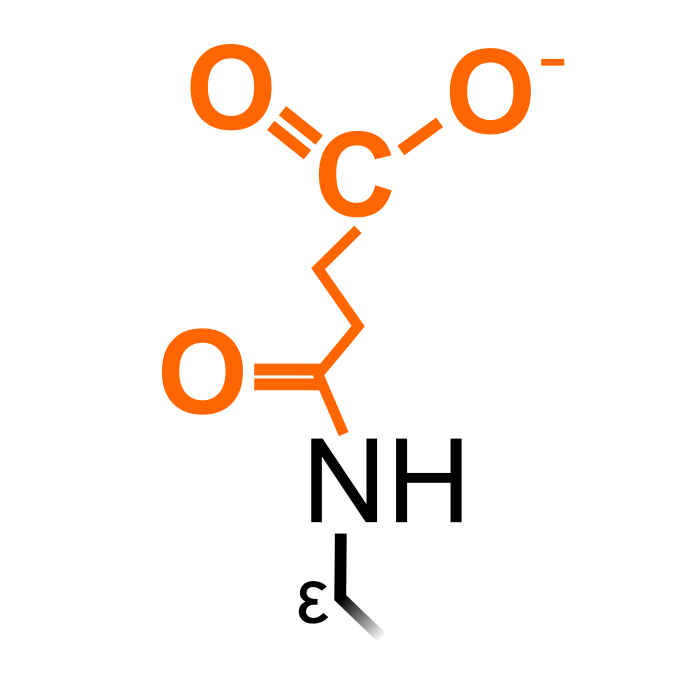
Succinylation is similar to acetylation and is induced by succinyl-CoA, though structurally larger and changing charge of Lys from +1 to -1 at physiological pH. These physicochemical properties are believed to have drastic consequences to protein structure and function, though further experimental studies are required for this PTM.
Δ Mass: +100.016 Da
sumo
SUMOylation
K
172
139
1
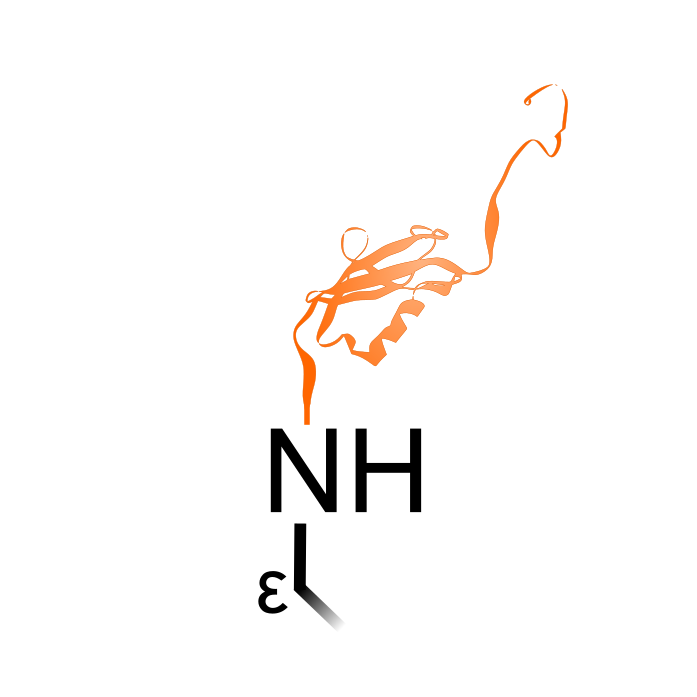
SUMOylation is the attachment of the Small Ubiquitin-like MOdifier (SUMO) to lysine sidechains of target proteins. SUMOylation can affect the protein function in diverse ways. Like ubiquitin, SUMOylation is also controlled by E1 activating, E2 conjugating and E3 ligating enzymes, and reversible due to deSUMOylation proteases. For the detection of SUMO sites, a mutagenized SUMO was engineered and expressed in Arabidopsis (Rytz et al. (2018) Plant Cell 30: 1077-1099). This substituted an Arg instead of His at the C-terminus to leave after trypsin cleavage a four amino acid peptide remnant Glu-Thr-Gly-Gly.
Δ Mass: +349.149 Da (GluThrGlyGly tryptic remnant)
ub
Ubiquitination
K, Rare: N-term
18038
8315
11
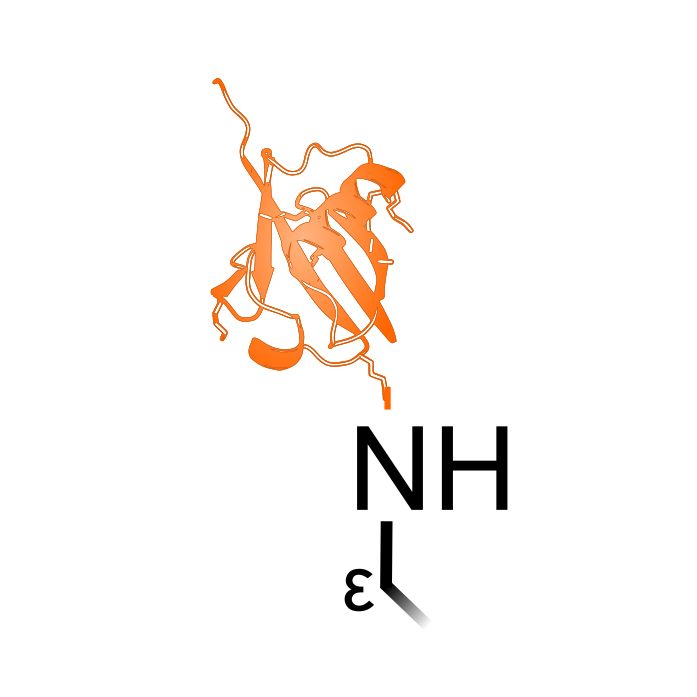
Lysine ubiquitination is the addition of the 8.5 kDa protein ubiquitine to Lys side-chains at the epsilon (ε) amino group. Ubiquitination involves three main steps: activation, conjugation, and ligation, steps all catalyzed by specialized enzymatic families. Either a single ubiquitin protein (monoubiquitination) can be attached or a chain of ubiquitin (polyubiquitination). Ubiquitin chains will target proteins for proteasomal degradation. Although classically associated with its degradation function, ubiquitination can also alter the protein cellular location, and steer protein interactions. Trypsin cleavage leaves a GlyGly peptide remnant that can be used to identify ubiquitination sites. Note that ubiquitination on the N-terminus is considered a separate PTM type 'N-terminal Ubiquitination'.
Δ Mass: +114.043 Da (GlyGly tryptic remnant)