|
|
|
|
|
|
||
| Home | ||
| Tutorial | ||
| Case Study | ||
|
|
Manual | |
| Data | ||
| General Info | ||
| Template Matching | ||
| Aligning Datasets | ||
| DTW algorithm | ||
| Thanks | ||
| Download | ||
| Manual | ||
Warping Modes GenTχWarper operates in two main modes: template matching mode and aligning datasets mode. The working mode must be selected at the start from the WarpMode menu.
At any time during the run of GenTχWarper the working mode can be changed. However, bear in mind that by switching from one mode to another you will loose the results obtained in the former one, unless you have saved them to a file before moving to another mode. 1) Template Matching Mode Interface
2) Aligning Datasets Mode Interface
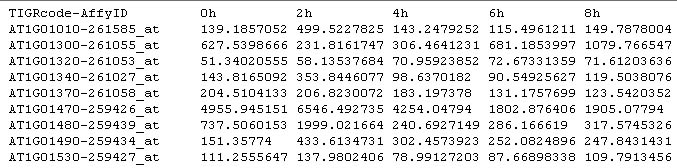
Input/Output GenTχWarper currently accepts as a valid input excel files and tab-delimited text files. The top row and the left most column of the input files are preserved for non-digital information, as for instance experiment and gene identifiers and anotations:
In both modes, the program can export the results to excel files or tab-delimited text files. The load, save, reset and exit functions are accessible via the File menu or via the tool bar buttons:
Data Adjustment In both modes the performance of the core alignment algorithm is subject to modification via several parameters: data adjustment, metric, offset, and anchor point. The data adjustment option enables z- and log2-transformations to be performed on the input expression profiles before alignment. They can be activated via the Data Adjustment menu or via the buttons:
The z-transformation can
be relevant when the general shape rather than the
individual gene expression amplitudes at the
different time points is important. The expression
value of each gene i at any time point
t is adjusted by substructing the mean and
dividing with the standard deviation of the
expression values of gene i over all the time
points. However,
this transformation should be used with caution,
bearing in mind that the expression levels of low
expressed genes will be amplified by it. The
log2-transformation involves normalization of the expression
value of each gene i at any time point
t with the expression value of the same gene
i at time point zero and then taking log2 of
the ratio. This transformation may be
essential for performing between experiment or
between spicies comparisions of gene expression time
series.
Metric The Metric menu allows a flexible choice between 3 different distance measures: Manhattan, Euclidean, Chebychev and Pearson correlation. Alternatively, the desired distance measure can be selected via the buttons:
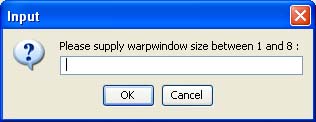
Warping Window The Warp Window menu enables the application of a warping window constraint during the calculation of the DTW distance. It leads to a reduction of the search space and consequently speeds up the processing of large microarray data sets. The warping window can alternatively be activated via the button:
The size of the warping window must be given in a pop-up window indicating the allowed lower and upper boundaries.
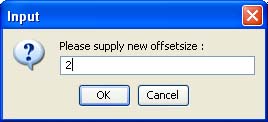
Any integer value within the specified range is accepted as a valid window size. For instance, a size 1 will mean that the optimal alignment path is allowed to deviate maximum one position to the left or to the right from the diagonal. The narrower the warping window is, the faster the final alignment will be produced. Additionally, the side effect of the application of a narrow warping window is that the optimal alignment path will be forced to stay close to the diagonal. The extreme usage of the warping window feature however, may have a negative effect on the accuracy of the alignment. Offset The Offset menu enables specifing a non-zero value as an offset parameter. This allows to slide the sets of expression profiles against each other along the time axis and in this way discovering genes with similar trajectories but shifted in time. For instance, putative targets of a known transcription factor can be identified by using its profile as a template in the template matching mode and evaluating the list of the best matching genes for different offset values. Additionally, the offset parameter can be useful in case the biological process under study displays a phase shift due to the design of the experiment. Note that the offset option can equivalently be accessed via the buttons:
The left button is on-off switch, while the right one allows setting a new offset value in the window below. Any integer value is accepted as a valid offset. For instance, an offset 2 will mean that the blue profiles are shifted with 2 time points to the right. Analogously, a negative offset will cause respective shift to the right of the red profiles.
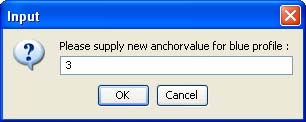
Anchor Point The anchor point option provides the possibility to explicitly fix together a pair of time points coming respectively from the two sets of expression profiles to be aligned. Setting an anchor point might be very useful in case there is detailed information about the exact times when the compared biological processes go through some fixed state. This feature is also useful when one of the time series to be compared is sampled during a shorter time interval than the other. Presently only one anchor point can be specified in GenTχWarper via the Anchor menu or alternatively via:
The left button is on-off switch, while the right one allows setting the coordinates of an anchor point in two sequential windows. In the first window, the index of the time point from the blue profiles to be anchored must be specified:
In the second one, the corresponding index for the red profiles needs to be supplied. Clearly, only positive integer values within the time index range of the time series are allowed as valid anchor point coordinates.
| ||

 .
. .
. .
. .
. .
.