| |
0. About the syntax
1. Defining components (mRNA/proteins)
+
comp
/
virtcomp
/
fixedcomp
2. Conditional processes: if ... then
+
if then
/
if
then transform
/
if
then block
/
if
then nonreg
-
Biology, mathematics
and the interface
-
The if-then statement
- Additivity
- Notes
- Note
about creation, block and break-down
3. Configuring the simulation
+
timepoints
/
starttime
/
const
/
sim
/ default
Appendix : Complete grammar
In the following sections, we specify each statement
(=recognized command) of the simulator. We will represent them as
"regular expressions".
Regular expressions are commonly used to define
the syntax of a line of text. They tell what is allowed so that
the simulator will understand a command. They usually are a keyword (the
command), followed by a number of optional arguments (=extra
information).
A
regular expression describes the real command as follows:
- 'text' means the literal text as it is typed between
the quotes;
- <text> means a number or a string that you can
freely choose (a string is
a sequence of letters/numbers/characters, starting with a letter);
- [abc] is zero or one occurences of abc;
- (abc)* is zero or more occurences of abc;
- (abc)+ is one or more occurences of abc;
- ( a | b ) means either a or b;
- text (italic font) will be a pointer to another
regular expression.
For example, the regular expression (
'bla' | 'a' )* would allow the text blabla and
ablaablaaaa , but not blabl .
Now we will give each statement's syntax along
with a few examples, and explain it a bit more detailed, in the following way:
|
statement's name
|
|
statement's syntax in the form of a regular expression
a few examples
|
|
comp |
'comp' <name> [<initialAmount> [<degradationRate>] ]
[colorDef | 'noplot']
|
colorDef |
<colorname>
| '(' <redValue0to255> [',']
<green> [','] <blue> ')'
red
green
light_blue |
(255, 0, 0)
(255 255 0)
(128 128 255) |
|
comp CDC25
comp CDC25 100
comp CDC25 100 0.05
|
comp CDC25 red
comp CDC25 100 red
comp CDC25 100 0.05 red
comp CDC25 100 0.05 (255, 0, 0) |
|
|
All components (mRNA or proteins) you wish to use in
a simulation must first be defined with the comp statement. This
statement can have up to four arguments: the component's name, an initial
amount, a degradation rate, and a color. The
component's initial amount
is how much component there is at the start-time defined with the
starttime command. If you don't explicitly define starttime,
the simulator will use the default value for the start time, which is
"time = 0". The optional
degradation rate (which must always be given as the third argument) reflects natural break-down
by proteases, nucleases etc. For example a degradation rate of
0.05 would destroy 5% of the present component per time unit.
Note that component units and time units are arbitrary. These
can be numbers of molecules, moles, seconds, days etc.
As a last argument, you can give a color. If you do
this the component will also be plotted in the combined view window
of SIM-plex, in the requested color. If you don't give this
color argument then the component won't appear in the combined view but
will only be plotted in the separate-views-window.
You can give a colorname from the X11 list, or
define it by its red, green and blue values (0=no color, 255=maximum of
that color), for example (255,0,0) is pure intense red.
Instead of a color, you can type noplot. This will make the plot
dissappear from the Combined Plot as well as from the Single Plot
window. This is useful when you want to focus on other components.
|
virtcomp |
'virtcomp' <name> =
[<factor> ['*'] ] <compName>
( '+' [<factor> ['*'] ] <compName> )*
[colorDef
| 'noplot']
virtcomp V = A + B
virtcomp V = 0.5 A + 0.8 B
virtcomp V = 0.5*A + 0.8*B
virtcomp V = 0.5*X + 0.8*Y + C |
virtcomp V = A + B
red
virtcomp V = 0.5 A + 0.8 B red
virtcomp V = 0.5*A + 0.8*B
red
virtcomp V = 0.5*X + 0.8*Y + C red |
|
The concept of a virtual component allows
the simulator to deal with problems like the
following: if a protein A is produced, and at the same time B is
produced through a different process (for example, it's a less
active derivative of A), and A and B have an additive effect on the
regulation of C. How would you express this in statements like "if ...>...
then ..."? You would need something like "if (...+...) > ...
then ...". To avoid additional complexity in the if-then definitions, we allowed defining virtual
components. In the example, the virtual component V
is defined as "A + 0.5 * B", and this
virtual V subsequently is the regulator of C. To be
able to handle this feature, the mathematics of Piecewise Linear Differential Equations
(PLDE) needed to be extended a little in SIM-plex. Next to the
piecewise linear differential equations, when defining virtual components,
also linear combinations of PLDEs are used. Note: you cannot define a
virtual component in recursive terms or with component names that are not
yet defined.
|
fixedcomp |
|
'fixedcomp' <name>
[ 'repeat' ] <timepoint> <compAmount>
( ',' [ 'repeat' ] ['add'] <timepoint>
<compAmount> )*
[ colorDef
| 'noplot' ]
(Note: only one
occurence of 'repeat' is
allowed )
fixedcomp comp1
0 0, 11 0,
13 10, 15 0,
31 0, 33 10,
35 0 blue
fixedcomp comp2
0 0, add 11 0,
add 2 10, add 2 -10,
add 5 0,
add 11 0, add 2 10,
add 2 -10, add 5
0
fixedcomp comp3
repeat 0 0, add 11
0, add 2 10,
add 2 -10, add 5
0 |
A fixed component represents a pre-defined
course of a component's amount through time. It can be used if you have
measured quantitative data for one component, and you want to see how
hypothetically dependent other components react.
The three examples clarify how the fixedcomp
statement works.
- The example with 'comp1' is the simplest: it defines a sequence of
timepoint+componentamount couples separated by comma's. If you try it
out in SIM-plex, you will see a profile with two peaks.
- The example of 'comp2' (two lines long!) defines the exact same profile
as for 'comp1', but now uses the add keyword, that adds a
time+amount to the last co-ordinates. This is useful because it makes a
profile a lot easier to change. And in the example you could e.g. use
copy-paste to quickly add another peak.
- The example of 'comp3' uses the repeat keyword to define an
infinite number of peaks like the two peaks in the 'comp1' and 'comp2'
profile. Note that the repeat keyword can occur anywhere between the
co-ordinates (though only once). This way you can first define a
start-profile, which is then repeatedly followed by the sequence after
the 'repeat' keyword. Note that the first component amount defined in
the repeat tail should be equal to the last amount, to allow the repeats
to link up.
|
if ... then ... |
'if'
condition ('and' condition)*
'then' <compName> <creationRate>
|
condition |
'true'
| ( <compName> ( '<' | '>' | '>=' ) (<threshold>) )
|
true |
(This is the way to define "always", or
"constitutively expressed") |
|
Rum1 < 40 |
(means: "if the Rum1 geneproduct is below
the threshold of 40") |
|
Cdc25 > 50 |
(means: "if the Cdc25 geneproduct is
above 50 units") |
|
if true then
MPF 5
(MPF could be the "mitotis-promoting factor" complex)
if Cdc25 > 50 then MPF 5
if Cdc25 > 50
and Rum1 < 40 then MPF 5 |
Biology, mathematics and the interface
If-then statements are at the heart of the simulator's
interface. They formulate what happens in a genetic regulatory network, in a
stepwise way. They approximate gene-activation or repression in a
step-wise manner, as if it happened with switches, but then allowing the
possibility to include more than one step between the "on" and the "off" state.
Gene regulation may happen by several proteins
that bind to a gene's promotor sequence, together controlling how active
the gene is expressed. Generally, the relation that describes a
gene's (de/)activation in function of a regulatory protein's
concentration, usually is
sigmoidal (the mathematical function has
a sigmoidal shape(1).). In the
Piecewise Linear Differential Equations (PLDE) model, these sigmoids are approximated by step functions. In case
of activation, this means that as long as the regulatory protein's
concentration stays below a certain threshold, the gene is in "off"
state, and as soon as the regulatory protein reaches the threshold or
rises above it, the gene is in "on" state. This is an
approximation but Glass and Kaufmann showed(1)
that a biological switching network that is approximated in this way,
can have the same outcome as the original network.
The SIM-plex simulator is built on the PLDE model, and offers a
very user-friendly interface that lets the user define these kinds of
regulation without being bothered much by the underlying mathematics. The
goal of the interface is to let the user work at a level above the
mathematical equations, 'closer' to biology. When a simulation is executed, there
first is a
translation of the user's statements to mathematical equations. This
translation can happen very transparent because there is an exact
relation between the user's if-then statements and the stepwise
activation functions that the PLDE model uses.
The if-then statement
Most genes are active only under certain
conditions. For example: only if protein A is present in sufficient
abundance, and protein B is virtually absent, the transcription of C is
activated. This condition and the resulting gene-activation can be
formulated in SIM-plex as "if A > 20 and
B < 5 then C 3".
You can see that after the if keyword,
you can build a condition that is a chain of simpler conditions that
are separated by the and keyword. A simple condition always
compares the available amount of one component to a certain threshold.
After the then
keyword, you can give a component's name and the rate at which this
component should be created when the all of the defined conditions are
met.
Knowing an exact value for a threshold would of
course be the ideal case, but with the current status of biological
knowledge we often only know relative values, like "This
component is
very abundant. Some other component is less abundant. And some are not
present at all at some timepoint." Therefore it is not crucial
- at this point in time - to use exact values. Estimates are good
enough. As long as you see that the network you defined is able to show
the expected behaviour with a given set of parameters, it is a good
indicator that you know most of your network's components. If not, then
you can start introducing hypothetical components and see if the
simulator is able to predict functionality for this new network. In this way SIM-plex
can be a useful tool for hypothesis generation.
Additivity It happens a lot that
gene activation changes as soon as an additional protein comes along to bind
to the promotor. To facilitate the definition of such events, the if-then
statements in SIM-plex were designed to work additively.
At each point in time, all statements that create a component P
under a condition 'true' are summed together.
Take the example from the tutorial:
if A > 20 then B 3
if B > 20 then B 2
In this example, when A rises above 20, B is activated and created at a
rate of 3. After that, when B rises above 20, B enhances its own
transcription, by creating an additional 2 component-units per time-unit. From
then onward, the creation rate of B increases to (3 + 2 =) 5.
Notes
- In any if-then statement, also in the
variants that follow below, you can also give threshold values and
creation-rates by means of the name of a user-defined constant (see below
how to define constants).
- Keep in mind that there is a difference in meaning between positive
and negative creation rates. It was explained in the tutorial, and will
be repeated shortly (under the explanation of the if ... then ... block statement).
- Component units and time units are arbitrary. These
can be numbers of molecules, concentrations, seconds, days etc.
- The underlying mathematical model works with half-open intervals.
Every border between two intervals (divided by a (de/)activation
threshold), always belongs to the upper interval. For example if the
real numbers are divided by two thresholds, 4 and 8, then there will be
piecewise-linear-differential-equations defined over each of the three
intervals: [0, 4[ and [4, 8[ and [8, +∞[.
That is why conditions are only built with the comparators <, >, and
>=. (Note that >= means ≥).
( > will have the same mathematical translation as
>=. It was only made available for who wants to type it
absolutely correct).
Note that [a, b[ means "everything between a and b, a
inclusive, b exclusive".
- Because of Note 4, the special
condition "X > 0" will always yield "true" (because > equals >=, and
the first halfopen interval is [0, firstThreshold[.
Because we still wanted it to be possible to make a distinction
between quasi-zero and more than this 'quasi-zero', we addressed
this problem by making SIM-plex translate any condition "X >
0" to "X > lowNonZeroThreshold". This
lowNonZeroThreshold is a very low
threshold that has a default but adjustable value in SIM-plex (see
below, under the default section).
|
if ... then ... block
... |
|
'if'
condition ('and' condition)*
'then' <compName>
'block' [ ( <blockFactor0to1>
| <additiveNegativeBlock> ) ]
if Rum1 > 40 then MPF
block
(complete block)
if Rum1 > 40 then MPF block
0.15
(partial block, 15% gets through)
if Rum1 > 40 then MPF block
-5
(partial block, subtracts 5 units / timeunit) |
Even when sufficient transcription activating proteins
are binding to its promotor, a gene can be partly or completely
deactived if an inhibitor protein gets a grip on the promotor too,
thereby interfering with the recruiting of the transcription machinery. This is what a block
tail in an if-then statement can model.
There are three ways to use the if-then-block
statement:
| -
"... then X block" |
(without an extra argument):
this will completely block all creation of X, described by all
other if-then statements. |
| -
"... then X block addition" |
(with a negative addition):
this subtracts a value from the original creation rate, but
cannot make it lower than zero. |
| -
"... then X block factor" |
(with a factor between 0
and 1): multiplies the creation rate (áfter possible negative
additions), with the given factor.
For example "block 0.15" would restrict the creation rate to 15%
of its original value. |
To dertermine the type of block (multiplicative or
subtractive), SIM-plex makes a simple distinction based on the value
that follows the "block" keyword:
- if it is between 0 and 1, it's interpreted as a multiplicative
block;
- if it is < 0, it's interpreted as an additive block. (Adds a
negative rate). A few examples:
block 0.9
lets 90% of the transcription go on;
block 0
blocks everything, just like "block" without
argument;
block -2
subtracts 2 (or "adds
-2", hence "additive block") units per time-unit.
Note: in order to completely block the transcription of a network component,
the only valid way is with these "block"
statements. This will prevent context-dependency, like using +5, and -5
to get back to 0: the +5 could for example become a +8 at some time.
Thus using
"block"
is the effective way to be sure that creation really stops.
Naturally, the "block" statement cuts off
transcription-product creation rates to 0 when additive blocks sum together
to a negative rate. While additive blocks are only meant as extra
functionality, we believe that multiplicative blocks (with a factor) will be used in most cases.
|
if ... then ... nonreg ... |
|
'if'
condition ('and' condition)*
'then' <compName>
'nonreg' <creationRate>
if Cdc25p > 40 then MPF
nonreg 2.5 |
The difference between "... then A 5"
and "... then A nonreg 5" is that
the former models transcriptional creation which can be regulated by block statements, and
the latter,
"nonreg"-creation, can not be regulated by blocks.
To explain this, assume that one simulates supposing
that mRNA is very quickly translated to proteins, and only a single
"geneproduct" is used (instead of two, mRNA and protein).
When some biochemical operation happens so that the protein is produced
(e.g. dephosphorylation of its phosphorylated counterpart), then this
biochemical production part can not be affected by blocks, while the
transcription part can still be under a "block" at the same time.
Instead of using the "nonreg" tail it is preferred to use the more powerful if-then-transform statement,
which is described
below and it makes use of the nonreg-functionality.
Note that "nonreg" followed by a
negative rate has the same effect as when the "nonreg" would be
omitted (e.g. "A nonreg
-5" and "A
-5"), because break-down always
has a not-transcriptional cause, and can not be regulated by "block"
statements.
|
if ... then transform ... |
|
'if'
condition ('and' condition)* 'then'
'transform' (<sourceName> | '('
(<sourceName>)+ ')' )
'to'
(<targetName> | '('
(<targetName> )+ ')' )
<rate> [<lowestThreshold>]
if A > 40 then transform
B to Bphosph 2.5
if true then transform (A B) to C
2
if true then transform (A A B) to C
2
if true then transform (A B) to
(C D)
2
if true then transform (A B) to
(C D) 2 0.001 |
The section above shows an if-then statement with a transform
clause in its tail, which is actually a shorthand for:
"under the given conditions, break-down the source component", and
"under the given conditions, create the target component at the same
rate".
Thus the transform clause effectively specifies the two commands
together. Prior to simulating the interactions, this if-then-transform statement is translated into two
(or more) normal if-then statements like the above.
→ Note that special care had to be taken to let the creation of the
target stop as soon as the source is depleted. Therefore the condition of
the original transform statement is extended during the translation to
plain if-then statements, by adding the condition "and
sourceComponent > 0".
But this special condition gives problems with the mathematical
model that is used!
As explained under the plain if-then statement's "Note 5" and "Note 4", the
integration interval will be the half-open interval [0, firstThreshold[
. As this condition is always true (since >0 is also translated as >=0, see
also Note 4), this extra condition had to be changed into "and sourceComponent > lowNonZeroThreshold".
This lowNonZeroThreshold is a very low
default threshold that is adjustable in SIM-plex (see
below, under the default section). It can also be given as an
additional argument to the transform statement.
The following example shows how this translation happens:
"if A>40 then transform X to Y
5"
is translated to the following two statements:
"if A>40 and X > lowNonZeroThreshold then
nonreg
X -5"
"if A>40 and X > lowNonZeroThreshold then
nonreg
Y 5"
Note that "nonreg" is used because biochemical transformation
cannot regulated by block-statements, as described before. More general, an if-then-transform statement could
be used to model the binding of proteins, or the splitting of a
component, or for catalysing many to many. The "to"
makes the distinction between the "source" and "sink" side, and if one
side contains more than one component, it must be enclosed between
brackets. Note that when a reaction would need one A
and two B's to form C, then you could define:
"if ... then transform (A B B) to C <rate>".
--- We are aware that these not-regulatory
biochemical operations are quantitatively less correct in the mathematical
framework we use (i.e. linear creation/consumption and
exponential degradation). But in a genetic network that is
already a quantitative approximation of reality, we deem that this
method can still provide a very useable functionality to complete the
definition of these switch-like networks --- Note: you really should define a protein and its
phosphorylated form as two different components, because they are two
different biochemical entities too, which can be transformed into one
another.
Note about
creation,
block and break-down The if-then statement can have different tails,
with different meanings. These were already described above. Here is an overview:
|
•(1) |
|
"... then
A 5" |
= |
transcriptional creation; |
|
•(2) |
|
"... then
A block 0.15" |
= |
blocking the transcriptional creation; |
| |
|
|
|
|
|
•(3) |
|
"...
then
A nonreg 5" |
= |
non-regulated (unblockable)
creation; |
|
•(4) |
|
"...
then
A -5" |
= |
consumption of available component. |
(1) and (2) model transcriptional creation,
regulable by "block" statements, while (3) and (4) model
(non-regulable) biochemical creation and break-down.
Note that in (4), you could also add
"nonreg", but the negative rate already shows that it models
biochemical
breakdown, so it's not really necessary to tell this twice.
The following figure illustrates the sequence of
calculations that determines the final creation rate or break-down rate
in SIM-plex, in the most complicated case:
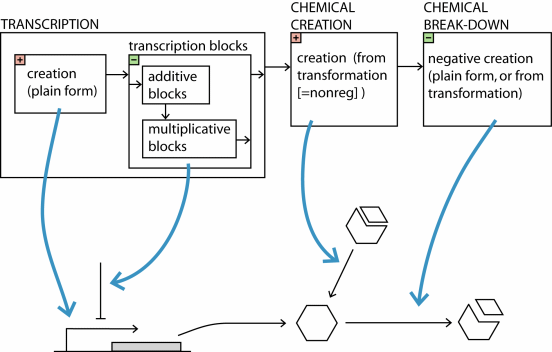
|
timepoints |
'timepoints'
oneTimepoints_def ( ',' oneTimepoints_def)*
|
oneTimepoints_def |
|
<timepoint> |
<beginTime> 'to' <endTime> [ 'step' <stepsize> | 'steps'
<nrofsteps> ]
2
0 to 100
0 to 100 step 0.1
0 to 100 steps 1000 |
timepoints 0
to 100
timepoints 0 to 200 step 0.1
timepoints 0 to 100 steps 1000, 100 to 200
step 0.1, 201, 202, 203
timepoints 200 to 300 |
With this command you can tell the simulator for
which timepoints you want to see the state of the simulated network. You
can give it in a list separated by comma's, and most easily with the
"step" or "steps" clause.
For example, "timepoints 0 to 10 steps 100"
specifies that you want 100 small steps between 0 and 10.
Consequently, you will get the simulation result for the 101 timepoints:
0, 0.1, 0.2, ..., 9.9, 10.
The command "timepoints 0 to 10 step 0.1" tells
that you want to make steps of size 0.1, also resulting in the 101
time points 0, 0.1, 0.2, ..., 9.9,
10. If no stepsize or number-of-steps is given, a
default number-of-steps is used: see below at the default
statement. The more timepoints you request,
the more detailed the simulation plots will be. This command is also
useful if you only need certain fixed timepoints, for example when you
work with the command-line interface.
|
starttime |
|
'starttime'
<time>
starttime 0
starttime 100 |
This statement determines at what time the simulation starts.
This is the time at which all the components have their 'initial amounts'.
It is not necessarily the first timepoint for which results will be
plotted: that should be done with the timepoints statement.
SIM-plex will report an error if the first
requested time point comes before this starttime, because backward
simulation is not possible.
|
const |
|
'const'
<name> '=' value ( ',' <name '=' value )*
const MPFthreshold = 30
const MPFthreshold = 30, Rum1_thr = 40 |
With this command you define constant values that
you can use for example for thresholds and rates in if-then statements.
|
sim |
|
'sim'
( 'bruteforce';
<timestep> | ... )
sim bruteforce 0.001
sim bruteforce 0.005 |
This command is intended to allow you to select which simulator you
want to use and what options you select for it.
Currently, however, only a "bruteforce" simulator has been
implemented. It performs the simulation in small timesteps. At each
timepoint, the creation/break-down rate of each defined component is
calculated, and a small integration step is executed. This integration
step is the "timestep" that you have to give as an extra argument after
the "bruteforce" keyword.
If you don't define a simulator, SIM-plex
always uses the "bruteforce" simulator as default, with a default
timestep as explained under default
statement.
Other uses of this command are reserved for the
future. Another simulator could be plugged in: one that jumps from threshold
to threshold in an N-dimensional space, which would in principle make
the integration even faster. The big difficulty with this approach is
what to do on so called 'black walls' and 'white walls', where solutions
are not unique: see (1).
The problem concerning what happens in the undefined threshold-zones, is also
the reason why the bruteforce-approach works with half-open intervals
over which the simulation happens (see above: Note 4). This way, there are no undefined
points.
(1)
Filippov AF. Differential Equations with
Discontinuous Righthand Sides. Kluwer Academic Publishers. (1988).
|
default |
|
'default' <defaultName> '=' <value>
(',' <defaultName> '=' <value>)*
default bruteforceSimStep = 0.001
default degradationRate = 0.05
default nrofTimepointSteps = 1000
default startComponentAmount = 0
default startTime = 0
default lowNonZeroThreshold = 0.1 |
This statement sets a number of default values
used by SIM-plex, to another value. In the examples above,
you see what SIM-plex uses as
default value for these defaults. To make
life easier, SIM-plex accepts a number of synonyms for each default
name. Also, the names are case-insensitive, which means: degrrate
= degrRate = DEGRRATE. SIM-plex
knows the following defaults (below the name are the
synonyms):
simStep
bruteforceSimStep
bfSimStep |
The timestep for the bruteforce
simulator. |
degrRate
degradationRate |
All the "comp" component
definitions that follow the redefinition of this default, and
that don't have an explicit degradation rate will get this value
for the degradation rate. |
nrofTimepointSteps
nrofTpSteps |
For people who don't give the "step" or
"steps" tail in the "timepoints" command. |
startComponentAmount
startCompAmount
initCompAmount
initComponentAmount
initialComponentAmount |
All the "comp" component definitions that
follow the redefinition of this default, and that don't have an
explicit initial component amount will get this value for the
initial component amount. |
| startTime |
For who doesn't use the starttime
command. |
lowNonZeroThreshold
lowThresh |
For an explanation, see Note 5
under the plain if-then statement, and also the explanation of
the if-then-transform statement.
→ This default should be
an order of magnitude higher than the bruteforce-simulator's timestep,
to make sure that the simulator does not, in one step, break-down more
of a certain component than there is available. |
An extra check was incorporated in SIM-plex
to make sure that "default"-definitions always happen
before all if-then statements; an exception is thrown if necessary.
For who wants more than the above, sometimes more
intuitive and user-friendly version of the syntax we next present the complete syntax of a network definition. Note: the colornames were
taken from the
X11 color names list.
statementList := (defaultReset |
declaration | systemEq |
simulationSpec)*
defaultReset := 'default' defaultAssignment (','
defaultAssignment)*
defaultAssignment := <defaultName> ['='] <value>
declaration := componentDef | virtCompDef |
fixedCompDef | constantDef
componentDef := 'comp' <componentName>
[<initialAmount> [<degradationRate>] ]
[colorDef | 'noplot']
colorDef := colorName |
( '(' <red0to255> [','] <green0to255> [',']
<blue0to255> ')' )
colorName :=
'black'|'blue'|'cyan'|'darkgray'|'darkgrey'|'gray'|'grey'|
'green'|'lightgray'|'lightgrey'|'magenta'|'orange'|'pink'|
'red'|'white'|'yellow'| etc. (full
list)
virtCompDef := 'virtcomp' <virtCompName> '='
[<factor> ['*']] <compName>
( '+' [<factor> ['*']] <compName> )*
[colorDef | 'noplot']
fixedCompDef = 'fixedcomp' <fixedCompName>
( fixedCompPoint (',' ['add'] fixedCompPoint)*
[',' 'repeat' ['add']
fixedCompPoint
(',' ['add'] fixedCompPoint)* ]
)
| ('repeat' fixedCompPoint
(',' ['add'] fixedCompPoint)*
)
[colorDef | 'noplot']
fixedCompPoint = timepoint compAmount
constantDef := 'const' <constantName> '=' <constantValue>
( ','
<constantName>
'=' <constantValue> )*
systemEq := 'if' totalCondition 'then'
(transformComponent |
formComponent)
totalCondition := simpleCondition ('and' simpleCondition)*
simpleCondition := bracketlessCondition |
( '('
bracketlessCondition ')' )
bracketlessCondition := 'true' |
( <componentName> ( '<' | '>' ['='] )
(<componentAmount> | definedConstant) )
definedConstant := <constantName> | '-' <constantName>
transformComponent := 'transform'
( <sourceComponentName> | '('
(<sourceComponentName>)+ ')' )
'to' (<targetComponentName> | '(' (<targetComponentName>)+ ')' )
(<rate> | definedConstant)
[ <lowestThreshold> | definedConstant ]
formComponent := <componentName> (
(<rate> | definedConstant)
| 'nonreg' (<rate> | definedConstant)
| 'block' [ ( <negativeRate> | definedConstant )
| (<factorBetween0and1> | definedConstant ) ] )
simulationSpec := starttimeDef | timepointsDef | simDef
starttimeDef := 'starttime' <time>
timepointsDef := 'timepoints' oneTimePoint_def
(','
oneTimePoint_def)*
oneTimePoint_def :=
<timepoint> |
<beginTime> 'to' <endTime> 'step' <stepSize> |
<beginTime> 'to' <endTime> ['steps' <nrofSteps>]
simDef := 'sim' ['bruteforce'] <timestep>
(1)
Glass L and Kauffman
SA. The logical analysis of continuous, nonlinear biochemical
control networks. Journal of Theoretical Biology 39, 103-129
(1973). |
|