|
|
|
|
|
|
||

| Home | |
|
| Tutorial |
| Manual | |
| FAQ | |
| Thanks | |
| Download |
| Get started with some examples | |||||||||||||||||||||||||||||||
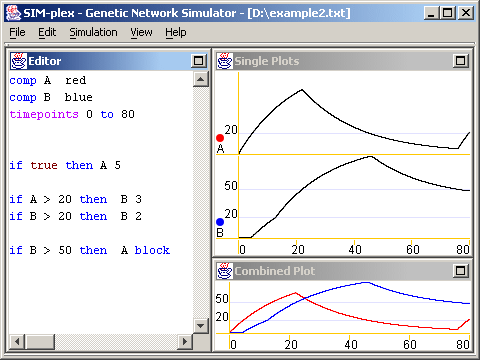
The simulator program shows three internal windows. In the image below, you see an example of its appearance after a very simple simulation.
You type the definition of a genetic regulatory network in the left window. After you choose Simulate from the Simulation menu, or simply hit F5, the right windows will show two different views of simulation results. The top window shows each geneproduct's amount through time separately, while the bottom window shows a combined view of a requested subset of the genes. The program can export the results to a file readable by Excel (tab-delimited), so while the right windows show results quickly, creating fancy graphs can happen outside the simulator. A quick introduction: (for more information, look on the Manual page)
You might wonder why the component amount of
A decreases in the above example. The reason is that this
reflects natural break-down by proteases, nucleases etc. The rate of
this degradation can be customized when you define the component with an
optional third argument.
There are three new things to learn from this example. (Note that it also illustrates comp's optional color argument that we already mentioned). First, it shows a statement with the "true" condition. Here it means "always create A at rate 5". Second, it shows that statements work additively.
At each point in time, all statements that create a component P and
have a condition that is true, are summed together. Third it illustrates the "block"
clause. In the example it means that all creation of A will be
blocked when the condition "B > 50" is true. Of course the degradation
(or break-down, see further) of component A already created, will
continue until this A is completely gone.
This illustrates one new statement: "if
... then transform
A to
B rate ". This can be used to model the
transformation of a protein A to its modified form (e.g. phosphorylated), here
called B. The slope in both graphs is again caused by proteolytic
or RNA degradation. In the combined view you can see that the slope nicely goes over
from the red to the blue graph. --- We are aware that these non-regulatory biochemical operations are quantitatively less correct in the mathematical framework that we use (i.e. linear creation/consumption and exponential degradation). However in a genetic network that is already a quantitative approximation of reality, we believe that SIM-plex still provides a very usable functionality to quickly and interactively define the global topology of switch-like networks --- Note: it is important to define a protein and its modified form as two different components, because they are two different biochemical entities, which can be transformed into one another.
This section is for those hungry for more.
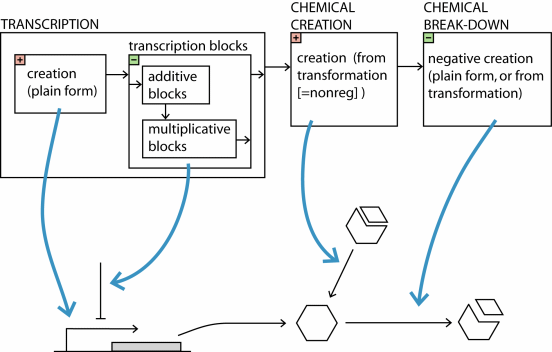
Note the difference between (2) and (4): the block-tail, and the "negative creation part". One may NOT use additivity like adding a +5 and a -5 to stop creation under certain conditions, because this would be context-sensitive! If the +5 would become a +8 at some time, or something else, then using "block" is the effective way to be sure that creation really stops. "Negative creation rates" can then of course still consume still available component. Also note the difference between (1) and (3):
transcriptional creation which can be regulated by block statements, and
"nonreg"-creation which can not be regulated by blocks. A schematic overview:
You are now ready to start simulating,
|