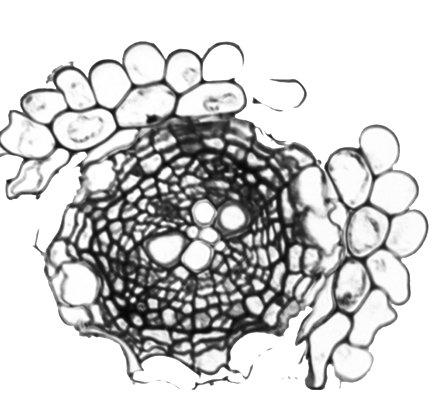
The vascular system of flowering plants develops from a handful of provascular initial cells in the early embryo into a whole range of different cell types in the mature plant. In order to account for such proliferation and to generate this kind of diversity, vascular tissue development relies on a large number of highly oriented cell divisions. The transcription factor complex formed by TARGET OF MONOPTEROS5 / LONESOME HIGHWAY (TMO5/LHW) is one of the major regulators of vascular proliferation and oriented cell divisions in the model plant Arabidopsis thaliana. The Vascular Development group studies plant vascular development, focusing on three major questions:
1) How is the actual selection of division plane orientation executed on a cellular level?
2) How is the TMO5/LHW pathway regulated and how does it interact with other developmental programs?
3) Which evolutionary innovations have enabled flowering plants to develop complex vascular systems?
1) How is the actual selection of division plane orientation executed on a cellular level?
2) How is the TMO5/LHW pathway regulated and how does it interact with other developmental programs?
3) Which evolutionary innovations have enabled flowering plants to develop complex vascular systems?
Cellular mechanisms of division orientation
As many cell files need to be formed during vascular development, it is crucial to understand how plants control the orientations of their cell divisions. Already in the nineteenth century, Errera and de Wildeman formulated rules stating that cells divide along the plane of the smallest surface that encloses a fixed cellular volume and is thus more commonly known as the ‘shortest wall’ rule. To deviate from the shortest wall rule, a cell must receive other cues in order to position its division plane differently. These cues can be, but are not limited to, developmental programs (e.g. lateral root formation or vascular development), genetic factors (e.g. transcription factors), responses to hormones (e.g. auxin or gibberellic acid) or environmental inputs (e.g. light or mechanical pressure). Nevertheless, it remains unknown how exactly these cues mechanistically determine the future division plane position and orientation.
Dr. Baojun Yang, funded by the FWO postdoc fellowship, addresses this question by identifying new components involved in division orientation with chemical genetics and imaging cytoskeleton dynamics using TMO5/LHW genetic tools.
Dr. Matouš Glanc, funded by an EMBO long-term fellowship, sees the orientation of cell division mainly as one of many manifestations of cell polarity. He aims to identify and characterize factors that connect inherent cell polarity cues with the cell division machinery and the TMO5/LHW pathway, focusing primarily on transmembrane proteins, which are his main area of expertise.
Dr. Baojun Yang, funded by the FWO postdoc fellowship, addresses this question by identifying new components involved in division orientation with chemical genetics and imaging cytoskeleton dynamics using TMO5/LHW genetic tools.
Dr. Matouš Glanc, funded by an EMBO long-term fellowship, sees the orientation of cell division mainly as one of many manifestations of cell polarity. He aims to identify and characterize factors that connect inherent cell polarity cues with the cell division machinery and the TMO5/LHW pathway, focusing primarily on transmembrane proteins, which are his main area of expertise.
Developmental control of vascular proliferation
Vascular tissue development depends (like most crucial developmental processes in plants) on large and complex regulatory networks, whereby genetic, hormonal and environmental cues must be integrated and interpreted by the appropriate developmental output. The group is currently working on several factors acting downstream of the TMO5/LHW complex, but we are only at the beginning of uncovering the whole regulatory network responsible for vascular proliferation.
Helena Arents, funded by PSB-VIB/Ugent, works on the functional characterization of novel players downstream of the TMO5/LHW heterodimer complex. She is looking into their possible spatiotemporal regulation within the TMO5/LHW pathway, following up on the legacy of the lab alumni Wouter Smet and Brecht Wybouw.
Max Minne, funded by a Ghent University BOF/GOA grant (which is a multidisciplinary collaboration with Prof. Tom Beeckman (VIB-UGent) and Prof. Tina Kyndt (UGent)), aims to map transcriptional responses during de novo organogenesis of vascular development, lateral root formation and nematode feeding site establishment at single cell resolution in Arabidopsis thaliana and Oryza sativa. Within this collaborative project, Max will focus on the role of cytokinin signalling during vascular development.
Helena Arents, funded by PSB-VIB/Ugent, works on the functional characterization of novel players downstream of the TMO5/LHW heterodimer complex. She is looking into their possible spatiotemporal regulation within the TMO5/LHW pathway, following up on the legacy of the lab alumni Wouter Smet and Brecht Wybouw.
Max Minne, funded by a Ghent University BOF/GOA grant (which is a multidisciplinary collaboration with Prof. Tom Beeckman (VIB-UGent) and Prof. Tina Kyndt (UGent)), aims to map transcriptional responses during de novo organogenesis of vascular development, lateral root formation and nematode feeding site establishment at single cell resolution in Arabidopsis thaliana and Oryza sativa. Within this collaborative project, Max will focus on the role of cytokinin signalling during vascular development.
Evolution of vascular tissues
Acquisition of vascular tissues is thought to be one of the main innovations that allowed plants to colonize dry habitats and grow beyond the centimetre scale. Still, our knowledge of the specific steps that allowed the transition from simple conductive tissues found in early land plants into complex vascular tissues remains limited. Our goal is to discover the molecular mechanisms that initiate vascular development. We have started to address this question in collaboration with prof. Dolf Weijers at Wageningen University by showing that both TMO5 and LHW are present throughout the plant kingdom. Yet, their heterodimerization which is crucial for vascular development, only appeared as a result of LHW neo-functionalization in flowering plants. Since we expect that multiple factors are involved, it is essential to be able to identify other developmental regulators in order to fully understand vascular development.
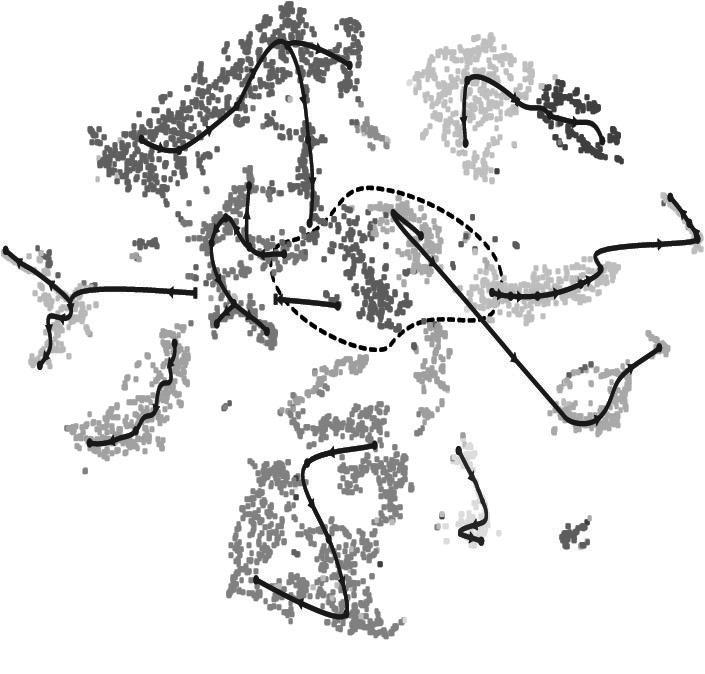
Jim Renema, funded by an FWO Odysseus grant, aims to discover novel factors involved in vascular tissue development. He is generating single cell expression profiles of multiple plant species. By comparing these expression profiles he will determine and characterize potential regulators.
Jim Renema, funded by an FWO Odysseus grant, aims to discover novel factors involved in vascular tissue development. He is generating single cell expression profiles of multiple plant species. By comparing these expression profiles he will determine and characterize potential regulators.
Single Cell Transcriptomics
The transcriptome of each cell practically serves as a molecular fingerprint as it is unique and bundles all information that define cell identity, function, physiological status or sensitivity towards its environment. Single cell transcriptomics comprises the isolation and examination of transcriptomes of single cells using RNA-Sequencing, in situ hybridization and the analysis of chromatin accessibility. Information from single cell transcriptomes can be used for a range of research areas, including fundamental and applied plant biology. These include for example long-standing questions in evo-devo plant specialization and adaptation strategies to biotic and abiotic stimuli. The technology can be used to accelerate targeted plant breeding and genome engineering and precision fertilization. The Plant Single Cell platform at PSB offers the infrastructure to guide projects from the drawing board to comprehensive single cell datasets. Additionally, we continuously optimize protocols and de-risk novel single cell methods and technologies for various plant species. This also includes translating single cell research from model species to crops in the AgBio Single Cell Accelerator. Publicly available scRNA-seq datasets from our platform can be accessed via our on-line browser tool.
Dr. Carolin Seyfferth is hosted in the De Rybel lab as coordinator of the plant single cell accelerator program at VIB (launched in January 2020). The accelerator program aims to centralize single cell approaches and techniques at PSB and collaborating plant biotechnology companies. Together with VIB Tech Watch, the program provides a platform for knowledge transfer between academic and industrial institutes to join efforts in promoting single cell technologies in various plant species.
Dr. Carolin Seyfferth is hosted in the De Rybel lab as coordinator of the plant single cell accelerator program at VIB (launched in January 2020). The accelerator program aims to centralize single cell approaches and techniques at PSB and collaborating plant biotechnology companies. Together with VIB Tech Watch, the program provides a platform for knowledge transfer between academic and industrial institutes to join efforts in promoting single cell technologies in various plant species.