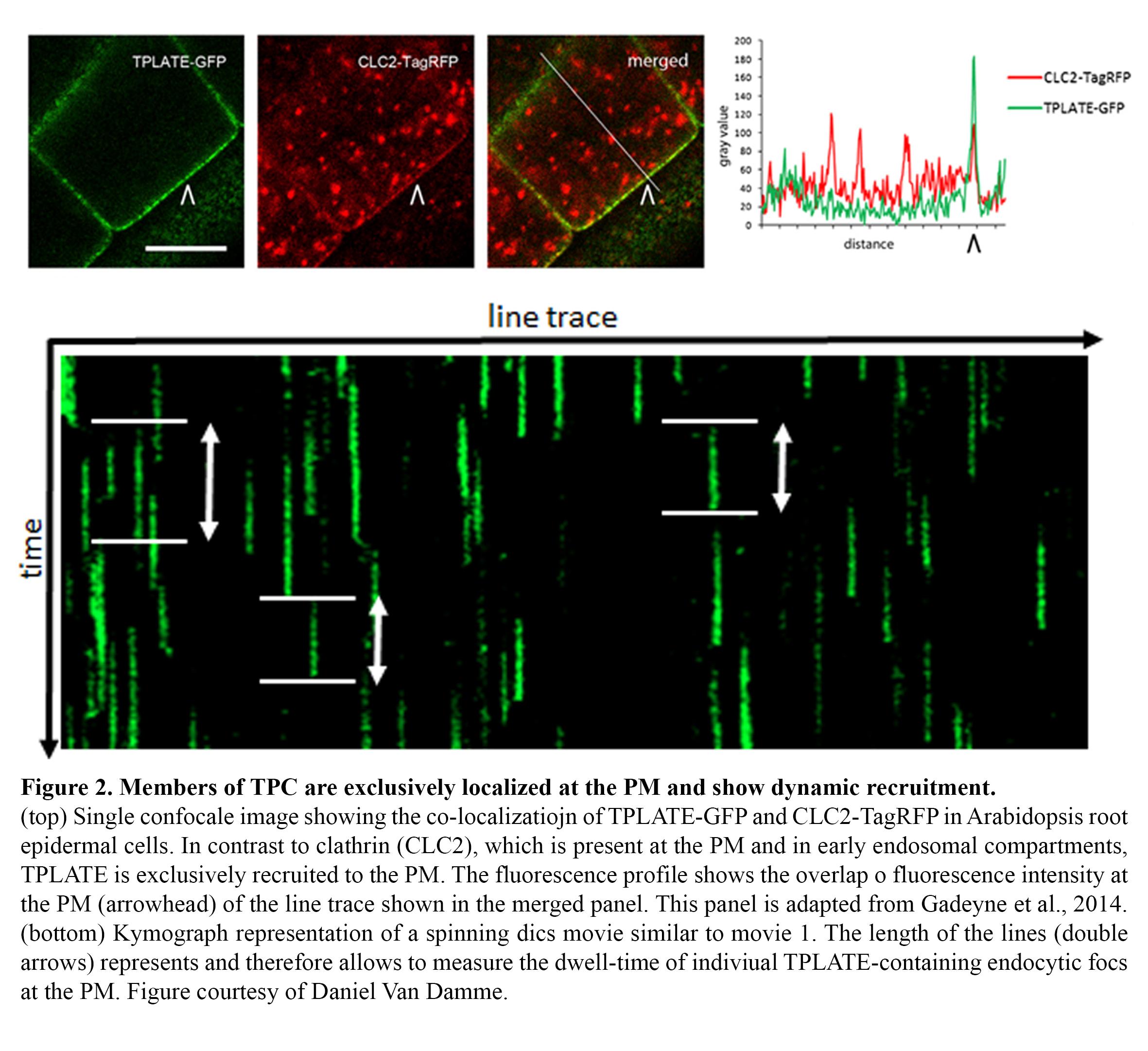
Endocytosis in plants operates differently compared to other model systems like animal cells or yeasts. TPLATE has been identified as a subunit of an octameric protein complex (The TPLATE complex, TPC) driving clathrin-mediated endocytosis at the plasma membrane in plant cells, working likely independently as well as in concert with the evolutionary conserved AP-2 heterotetrameric adaptor protein complex (Gadeyne et al., 2014). This complex represents an evolutionary ancient adaptor module which, although still present in Dictyostelium but not functional anymore, has been completely lost in the lineage leading to animal and yeast cells (Hirst et al., 2014). In contrast to other effectors of endocytosis, like clathrin, all TPC subunits localize exclusively to the plasma membrane and accumulate in highly dynamic endocytic foci, which show no lateral movement, allowing to accurately measuring the dwell-time of their recruitment via kymograph analysis (Figure 2).
movie
The movie shows an etiolated hypocotyl cell of a 4-day old functionally complemented tplate (-/-) Arabidopsis seedling. Dynamic TPLATE-GFP foci can be seen recruited and departing from the plasma membrane without lateral movement.The seedling was imaged with 500ms exposure time for 2minutes. The original images were processed with imageJ using a walking average of 4 frames and the movie plays at 20frames/sec.
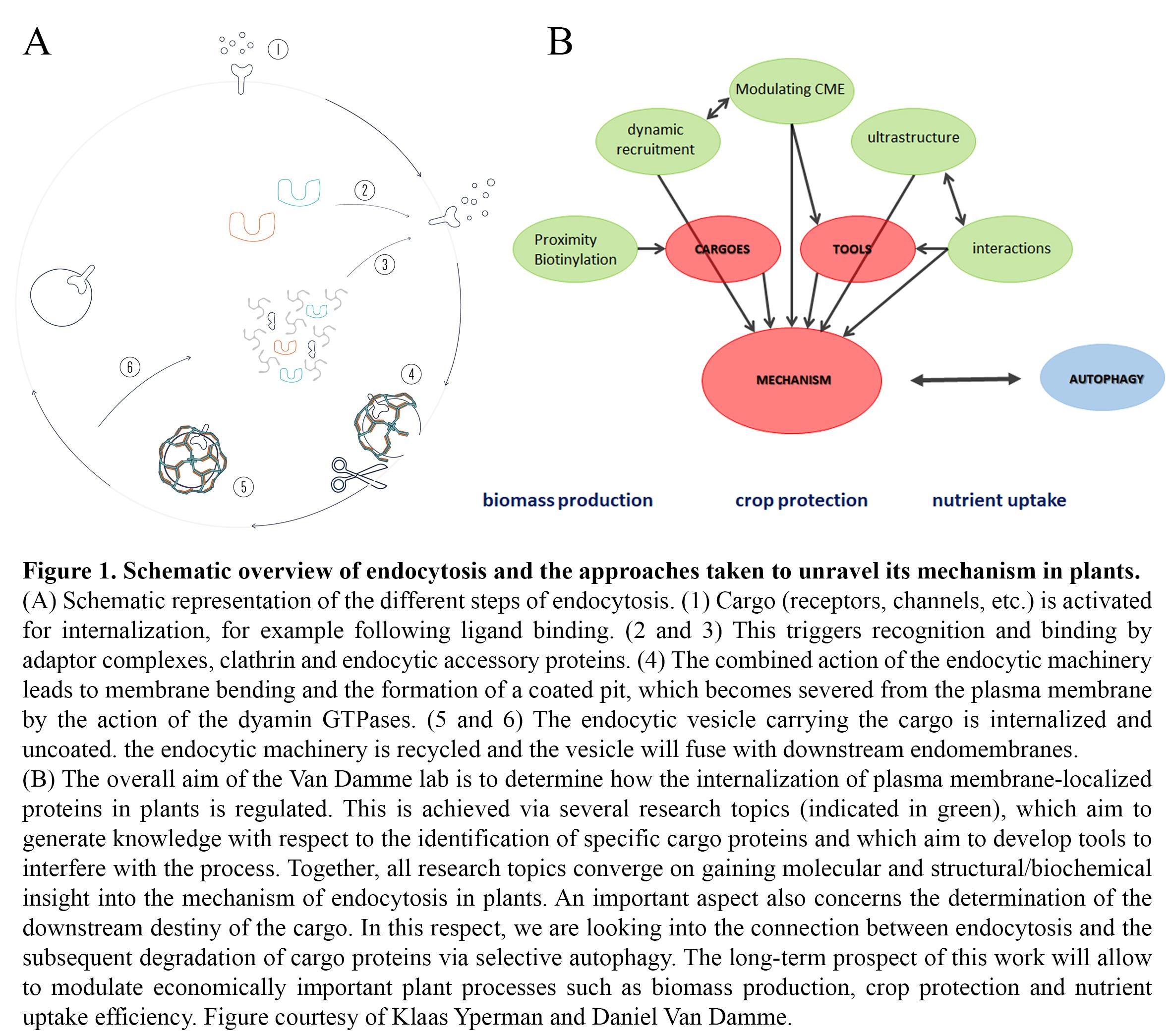
The overall objective of the research in the group is to gain a deep mechanistic insight into the developmentally essential process of clathrin-mediated endocytosis (CME) in plants, and how its regulation deviates from that in other systems. This will enable us to specifically modulate the abundance of plasma membrane proteins involved in nutrient uptake, toxin avoidance, cell wall formation but also hormone and defense responses. Understanding TPC-dependent CME will also provide insight into evolutionary aspects of endocytosis. In the framework of an ongoing ERC project, we are addressing the following questions with respect to unravelling the mechanism of endocytosis in plants:
- Is the evolutionary retention of the TPC in plants causal to specific cargo recognition?
- What are the temporal dynamics of TPC and CME effectors at the plasma membrane?
- How does acute removal of TPC subunits affect complex recruitment and CME?
- How is the TPC organized at the structural level?
- Which interactions occur and can we couple subunit/domain structures to functionality?
- How are endocytosis and autophagy-dependent degradation connected?
To answer these questions, we combine state-of-the art proteomics with highly dynamic multi-colour live cell imaging and structural biology, including protein crystallization and structural EM.
Proximity biotinylation uses a promiscuous biotin ligase, which causes biotinylation of proteins in the vicinity of the bait. These biotinylated proteins can be identified using mass spectrometry without the need to maintain the protein-protein interactions during the purification (Figure 3). This tool is therefore especially suited for interactions between cytosolic and transmembrane proteins. We are designing protocols to adequately perform proximity biotinylation in plants. This includes optimizing the temperature conditions, biotin concentration as well as the timing of biotin addition and the use of linker sequences between the ligase and the bait to achieve a bigger radius. We are also optimizing the detection of biotinylated peptides and we are developing methods to filter out false positives. This work runs in close collaboration with the functional interactomics group of Prof. Geert De Jaeger (PSB-VIB).
As endocytosis of transmembrane proteins (cargo) is initiated by the recognition of the cargo by adaptor proteins, elucidating the mechanisms that determine the interaction between the plant-specific TPC, the AP-2 complex and their cargo will allow modulation of cargo internalization. We aim to identify TPC and AP-2 complex cargo proteins by mass spectrometry analysis targeted biotinylation (bioID, Roux et al., 2012) using the third generation bioID, TurboID (Branon et al., 2018). Comparing interactors found using TurboID fused to AP-2 or TPC subunits will answer the question whether specific cargoes are recognized by these adaptor complexes.
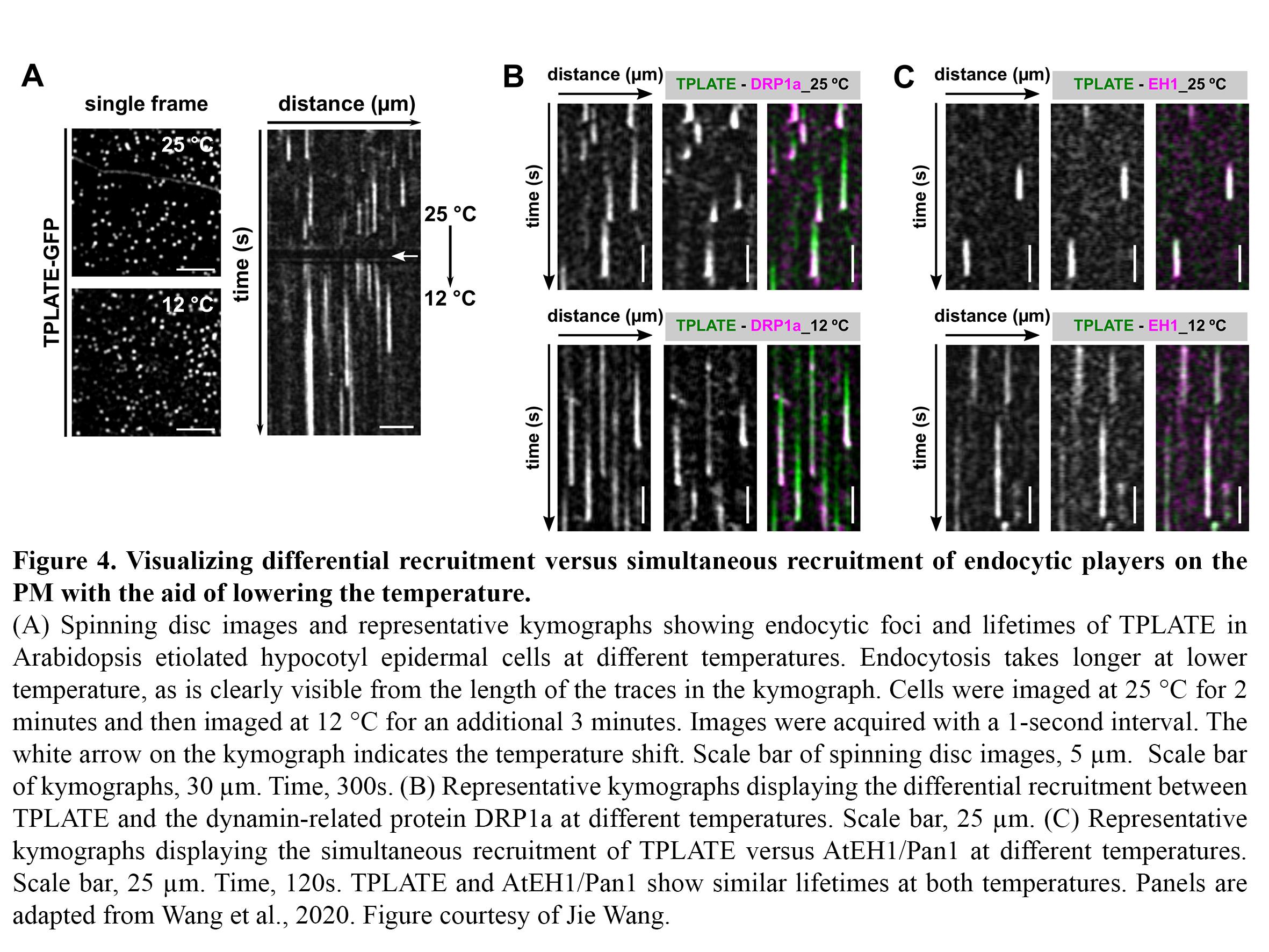
Dynamic imaging of fast-occurring processes is hampered by the fact that the signal/noise ratio of a fluorescent image is inversely related to the exposure time. The exposure time in turn limits the imaging speed and thereby the temporal resolution. We are increasing the temporal resolution of live-cell imaging plant samples by modulating the temperature of the samples in situ on the microscope stage as lowering the temperature slows down biological processes. Next to this, we use our temperature-modulatory setup also to study the effect of high temperature stress on plants.
CME is a fast and stepwise process and involves the sequential recruitment of several players. Adaptor complexes, the clathrin scaffold and endocytic accessory proteins all arrive and depart at different time points during the process. To visualize the temporal dynamics of the various players, we are generating fluorescently tagged functional fusions of TPC subunits and are performing dual colour spinning disc microscopy on complemented double mutant lines at ambient and at lowered temperature (Figure 4).
Many tools have been developed that allow to analyse loss-of-function mutants, including conditional silencing, genome editing etc. However, these tools often fall short for lethal genes that encode for highly stable proteins. In this case, conditional interference with gene function at the DNA or mRNA level does not result in an immediate effect and tools that interfere at the protein level directly are more efficient. So far, all tested knock-out alleles of TPC subunits cause fully penetrant male sterility and inducible silencing causes seedling lethality, albeit several days after induction (Van Damme et al., 2006; Gadeyne et al., 2014). Elucidating the mechanism of CME in plants requires tools to specifically interfere with the process. To avoid delayed effects due to the stability of the complex, we are developing strategies to interfere with TPC function at the protein level. We are exploring different paths to achieve this goal, ranging from rapamycin-dependent delocalization of the complex to the mitochondria (a.k.a. knocksideways, Robinson et al., 2010) to the development of temperature sensitive alleles (Figure 5). In collaboration with the nanobody core from the VIB, we have recently also started to explore the use of nanobodies directed against specific TPC subunit domains to interfere with protein function.
To generate novel insight into how vesicle trafficking is organized in plants, we use a multidisciplinary approach, combining live cell imaging with in-depth structural characterisation. We use E.coli as workhorse to express different domains of the endocytic TPLATE complex and in collaboration with A. Poterszman (IGMBC, Strasbourg) we are setting up expression systems in Sf9 cells for plant-specific multimeric complexes. Proteins are purified using classical protein purification techniques with the help of an ÄKTA system (GE Healthcare). Purified proteins are used to perform detailed biochemical characterisation experiments in combination with structural characterisation (X-ray crystallography - Prof. Savvas N. Savvides (IRC-VIB) and 4D-chains NMR - Dr. Kostantinos Tripsianes (CEITEC, Masaryk University, Czech Republic)).
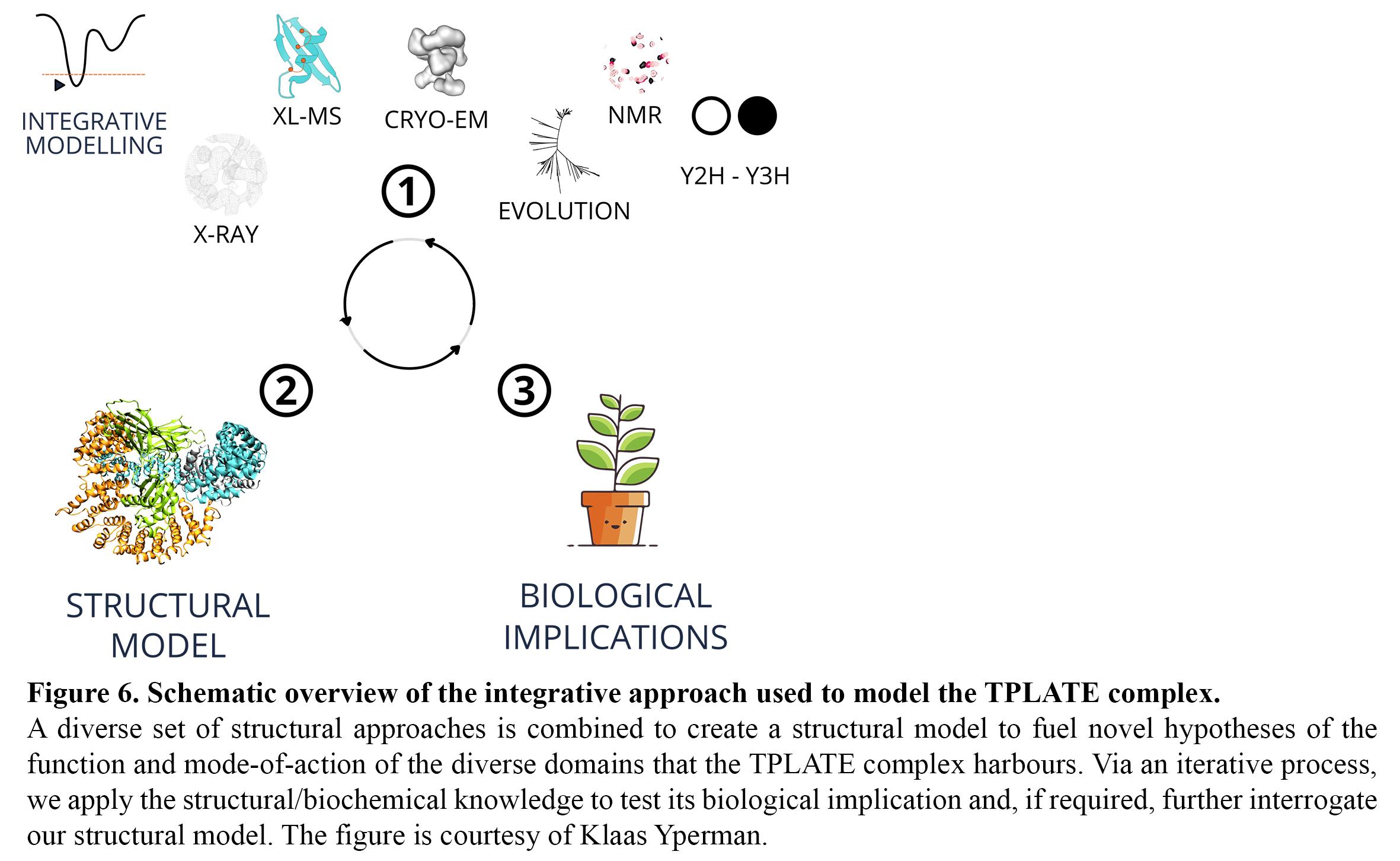
To obtain insight in multi-subunit complexes such as the octameric TPLATE complex as a whole, we purify native complexes via tandem affinity purification and ion exchange chromatography from Arabidopsis cell cultures. We use an integrative structural approach (Figure 6) that combines cross-linking mass spectrometry (BS3, DSSO) to identify inter-subunit crosslinks, single-particle electron microscopy (VUB-VIB Bio Electron Cryogenic Microscopy facility, Brussels), Y2H/Y3H and evolutionary insight in order to, via multiscale modelling, obtain the first ultrastructural insight of this adaptor complex.
The combination of our modelling approaches, XLMS, biochemical assays to determine for example lipid binding, domain structure determination and EM analysis will allow us to compare the structural organization of the TPLATE complex to other adaptor and coatomer complexes and to fuel novel hypotheses, which we subsequently test using live-cell imaging approaches.
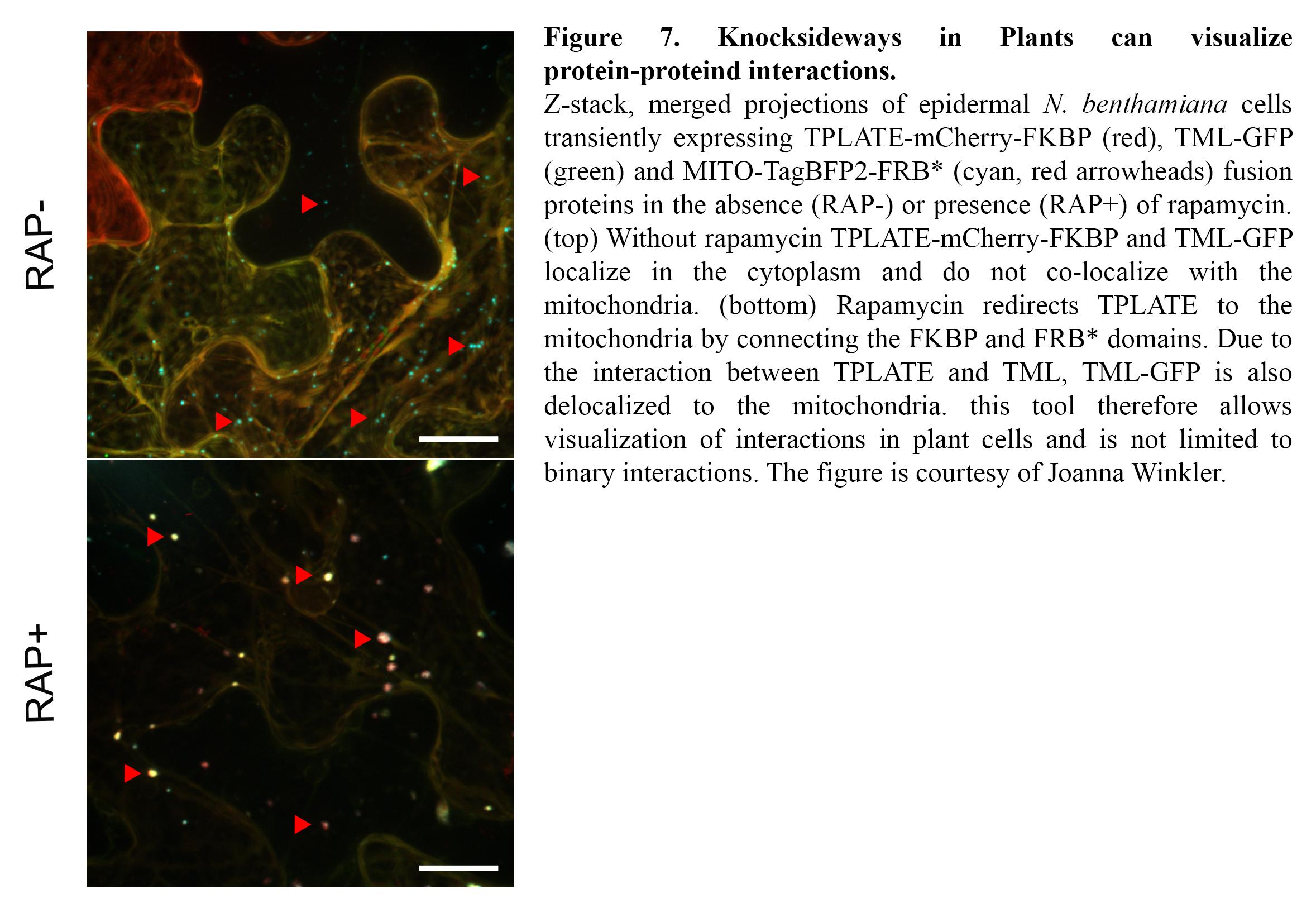
TPC is a very robust complex, yet our knowledge on the binary interactions occurring among its eight subunits is very scarce as standard interaction tests such as Y2H only yielded a single interacting protein pair (Gadeyne et al., 2014). To gain insight into the interactions, we are expanding our interaction assays to higher order interactions (e.g. Y3H) and we are developing imaging-based protein-protein interaction assays to visualize them. Knocksideways in Plants (KSP) for example can be compared to an intracellular Co-IP experiment. The tool uses the ability of rapamycin to alter the localization of a bait protein and its interactors via the heterodimerization of FKBP and FRB domains. KSP is inherently free from many limitations, which other PPI systems hold. It is an in vivo in planta tool, it is flexible concerning the orientation of protein tagging as long as this does not interfere with the interaction and it is compatible with a broad range of fluorophores. KSP is also a conditional tool and therefore does not require additional controls. (Figure 7).
Next to this, we are developing we are developing in vitro interaction assays (e.g. alphascreen PE) between recombinant protein domains to screen for chemicals that disrupt these interactions, and thereby impair TPC function. This work is done in close collaboration with the VIB compound screening facility.
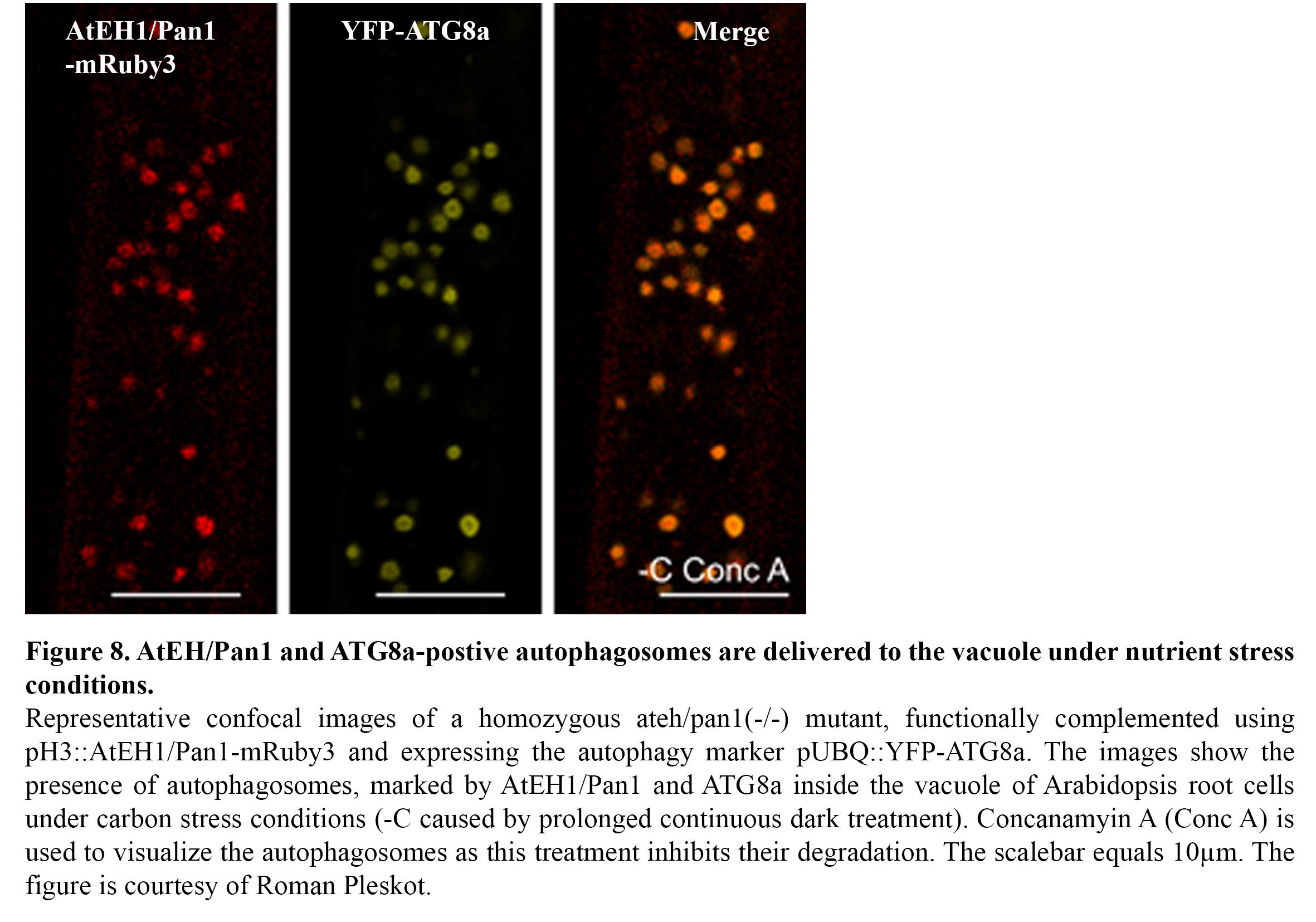
We recently discovered the existence of an autophagosomal degradation pathway, which operates between the ER-PM contact sites and the vacuole (Wang & Pleskot et al., 2019). In particular, two TPC members, AtEH1/Pan1 and AtEH2/Pan1 play an important role in this pathway (Figure 8) and several components of the endocytic machinery are associated with this pathway, either as machinery or as cargo. Ongoing research focusses on unravelling in detail how this pathway is initiated, as well as how it is connected to the membranes of the endomembrane system. Furthermore, we are exploring the role of this pathway under abiotic stress conditions such as elevated temperature and nutrient (C/N) stresses. The aim here is to gain insight how endocytosis and autophagy are interconnected during stress conditions. TPLATE for example was shown to be actively degraded under nutrient stress conditions (Wang & Pleskot et al., 2019).