The transcriptome of each cell practically serves as a molecular fingerprint as it is unique and bundles all information that define cell identity, function, physiological status or sensitivity towards its environment. Single cell transcriptomics comprises the isolation and examination of transcriptomes of single cells using RNA-Sequencing, in situ hybridization and the analysis of chromatin accessibility. Information from single cell transcriptomes can be used for a range of research areas, including fundamental and applied plant biology. These include for example long-standing questions in evo-devo plant specialization and adaptation strategies to biotic and abiotic stimuli. The technology can be used to accelerate targeted plant breeding and genome engineering and precision fertilization. The Plant Single Cell platform at PSB offers the infrastructure to guide projects from the drawing board to comprehensive single cell datasets. Additionally, we continuously optimize protocols and de-risk novel single cell methods and technologies for various plant species. This also includes translating single cell research from model species to crops in the AgBio Single Cell Accelerator.
PLANT scRNA-SEQUENCING PIPELINE
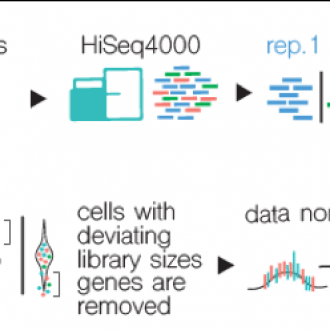
We established a comprehensive pipeline that enables high-throughput processing of scRNA-Sequencing samples from model plant species (e.g. Arabidopsis thaliana) and soon also crops (e.g. maize, rice). Sample processing, including cell isolation, cell enrichment, sequencing and analysis, is done entirely within VIB using available core facilities.
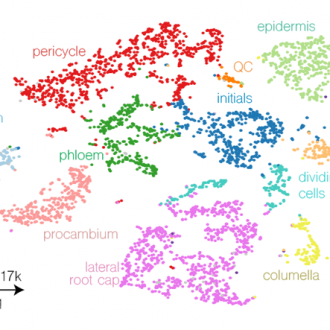
As an example of the current state of the art, a collaboration between the De Rybel and Saeys Lab generated a high-resolution single cell transcriptome map of the Arabidopsis primary root meristem, containing transcriptomes from over 5000 cells and an average of 6850 expressed transcripts/cell (Wendrich et al., 2020). Additionally, an advanced in-house bioinformatics pipeline for plant samples has also been established, allowing rapid and robust data processing, cell clustering, cluster annotation and visualization, and the analysis of developmental trajectories. A detailed visualisation of the transcriptome atlas and cell developmental trajectories is freely accessible under http://bioit3.irc.ugent.be/plant-sc-atlas/.
TECHNOLOGY IMPLEMENTATION
Advanced methods and technologies for single cell analyses develop very fast. We are continuously extending the portfolio of methods and technologies to study plant processes at a single cell level. Upcoming developments include:
- Advanced sample multiplexing: a method to boost cost efficiency and enable high-throughput screening.
- Single nuclei RNA-Sequencing: an alternative to single cell RNA-Sequencing to circumvent limitations that occur with the isolation of single cells.
- Spatial transcriptomics: adding spatial context to single cell transcriptomes.
Updates on technologies and methods that are established within VIB and the Plant Single Cell platform are regularly presented as part of the AgBio Single Cell Accelerator.
TECHNOLOGY BENCHMARKING
Early access to upcoming technology enables us to be on top of technological innovation and trends. Our current focus lays in benchmarking of technologies related to
- Automation of sample preparation
- Single cell RNA-Sequencing platforms
- Spatial sequencing platforms
Contact: The Plant Single Cell platform at PSB is operated by dr. Carolin Seyfferth and dr. Thomas Eekhout and hosted in the De Rybel Lab. Please contact Carolin and Thomas directly for more information.