We offer the plant research community and companies access to our latest interactomics tools, covering AP-MS and Turbo-ID based proximity labeling as complementary approaches.
Experiments are performed under Research Service Agreement, following well-established protocols mainly developed in Arabidopsis PSB-D cell suspension culture. As starting material, either cell cultures, callus tissue, seedlings or isolated plant tissues can be used. We have extensive experience with interactomics analyses in Arabidopsis1-4 and maize5,6. Moreover, our protocols have been successfully transferred for analyses in rice7, tomato2, medicago8 and canola. For efficient AP-MS experiments, we prefer to use Protein A/G-based tags. Alternatively, also AP-MS based on GFP tags can be requested. We have also ample experience with tandem affinity purification for the isolation of stable protein complexes at high purity.
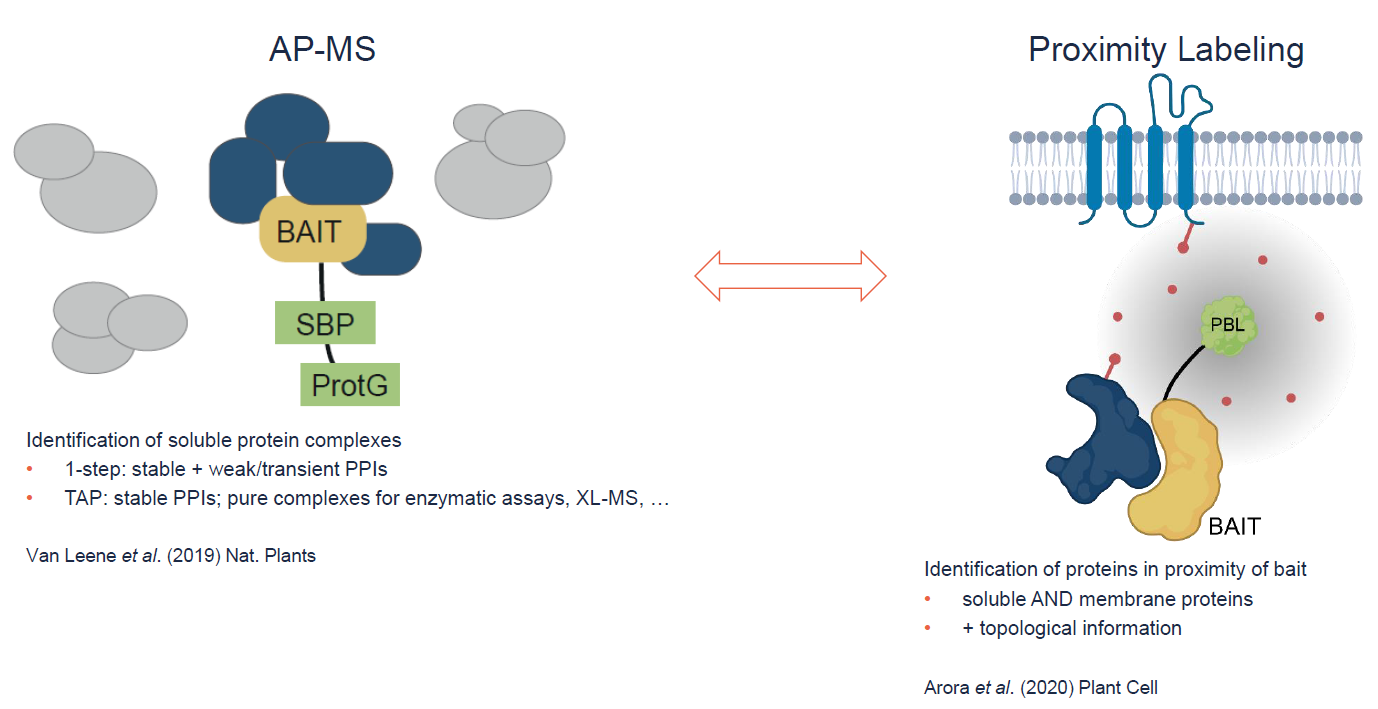
Schematic Overview of our AP-MS and proximity labeling technology (Figure by Van Leene and Yperman).
For identification of isolated proteins, we have access to state-of-the-art mass spectrometry instruments (Orbitrap Fusion Lumos and Q-Exactive HF). In order to pinpoint specific interactors, non-specific proteins are filtered out using a multitude of methods, integrating label-free quantitative MS tools and covering either a large dataset approach, control experiments or conservation of non-specific background proteins across plant species.
For cell cultures, we start from delivered bait clones, for experiments in plants, we start from delivered plant tissue. More details and conditions are provided after consulting us at dominique.eeckhout@psb.vib-ugent.be.
Information on our protocols can be found in following manuscripts:
- Van Leene, J. et al. Capturing the phosphorylation and protein interaction landscape of the plant TOR kinase. Nat. Plants 5, 316-327 (2019).
- Arora, D. et al. Establishment of proximity-dependent biotinylation approaches in different plant model systems. Plant Cell, 2020 Aug 25, online ahead of print.
- Van Leene, J. et al. An improved toolbox to unravel the plant cellular machinery by tandem affinity purification of Arabidopsis protein complexes. Nat. Protoc. 10, 169-187 (2015).
- Van Leene, J. et al. Isolation of transcription factor complexes from Arabidopsis cell suspension cultures by tandem affinity purification. Methods Mol. Biol. 754, 195-218 (2011).
- Besbrugge, N. et al. GSyellow, a multifaceted tag for functional protein analysis in monocot and dicot plants. Plant Physiol. 177, 447-464 (2018).
- Nelissen, H. et al. Dynamic changes in ANGUSTIFOLIA3 complex composition reveal a growth regulatory mechanism in the maize leaf. Plant Cell 27, 1605-1619 (2015).
- Dedecker, M. et al. Transferring an optimized TAP-toolbox for the isolation of protein complexes to a portfolio of rice tissues. Plant Mol. Biol. 91, 341-354 (2016).
- Goossens, J. et al. Isolation of protein complexes from the model legume Medicago truncatula by tandem affinity purification in hairy root cultures. Plant J. 88, 476-489 (2016)
Selection of published stories including AP-MS data generated in our research group:
- Houbaert, A. et al. POLAR-guided signalling complex assembly and localization drive asymmetric cell division. Nature 563, 574-578 (2018).
- Heyman, J. et al. The heterodimeric transcription factor complex ERF115-PAT1 grants regeneration competence. Nat. Plants 2, 16165 (2016).
- Gadeyne, A. et al. The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell 156, 691-704 (2014).
- Pauwels, L. et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788-791 (2010).