BiNGO
How to use GO annotation and ontology files provided by the GO consortium (www.geneontology.org)
BiNGO uses default GO annotations parsed from files available at NCBI (see version_information.txt in the BiNGO.jar file), but these are rather old and are being phased out. We recommend you use GO annotation files provided by the GO consortium (named 'gene_association.<provider>'), you can load these as custom annotation files in BiNGO. Just download the file from GO, select Custom it in the Select organism/annotation dropdown menu and choose the right file. Any file named gene_association. ... is handled as a GO Consortium annotation file by BiNGO.
The same goes for GO ontology files: download the file from GO, select Custom it in the Select ontology file dropdown menu and choose the right file. Any file ending on .obo is handled as a GO Consortium ontology file by BiNGO. If you choose an .obo file, the Select namespace panel in BiNGO becomes active. Use this panel to choose the (sub)ontology you want to use. If you select ‘---’, the full ontology is used.
You should be careful when using GO Consortium annotation files: only a few types of identifiers are supported, dependent on the organism (columns 2,3 and 11 in the gene_association files). Furthermore, the annotation files for some organisms are protein-centric, while others are gene-centric. The annotation file for Arabidopsis, for example, contains splice variants of genes, resulting in approx. 50,000 annotated entities, although there are only approx. 25,000 ORFs. If you do not want to include splice variants in your analysis, you'll have to use a custom reference set (list of return-separated identifiers) under ‘Select reference set’ or make a custom annotation, which can be done from within BiNGO (see below).
How to make your own annotation files
You can use your own annotation/ontology files in BiNGO. This feature is particularly useful if you want to :
-
•use gene identifiers other than those provided in BiNGO (e.g. Affymetrix probesets)
-
•use an annotation for an organism (e.g. Poplar) which is not included in BiNGO
-
•test your set of genes against a custom reference set, e.g. all the genes on a microarray or in a large-scale Y2H screen
-
•use ontologies other than those provided, e.g. KEGG.
The format of custom annotation/ontology files in BiNGO is the same as the Cytoscape annotation and ontology file formats. To make a custom annotation file, just parse your annotation into the following form :
(species=Saccharomyces cerevisiae)(type=Biological Process)(curator=GO)
YAL001C = 0006384
YAL002W = 0045324
YAL002W = 0045324
YAL003W = 0006414
YAL004W = 0000004
YAL005C = 0006616
YAL005C = 0006457
YAL005C = 0000060
YAL007C = 0006888
YAL008W = 0000004
...
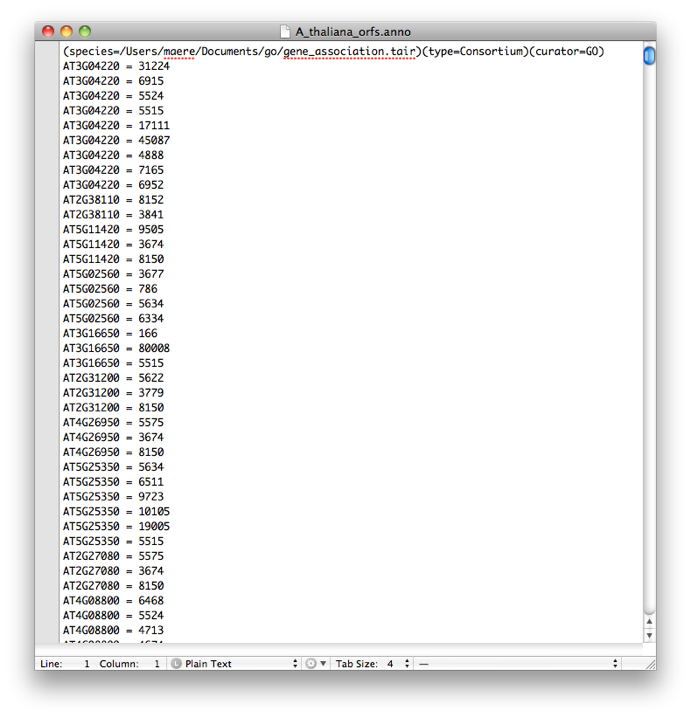
On each line, you link a gene identifier (before the equal sign) to an ontology category identifier (after the equal sign). The first line is obligatory and contains fields giving info about the species, the type of ontology and the curator.
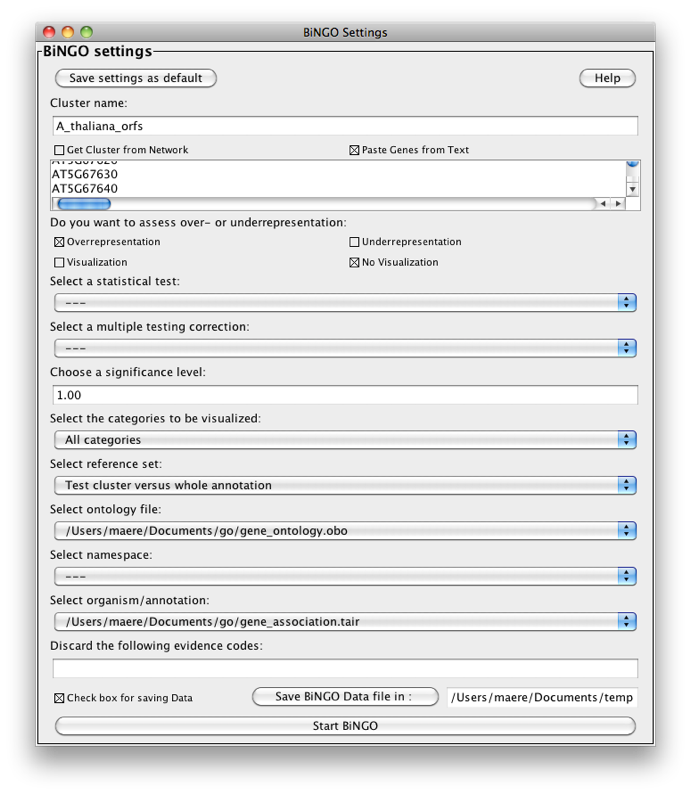
You can parse default annotation files into custom annotation files using BiNGO (provided the custom annotation you want is a subset of some default annotation). Suppose you want to make a custom annotation for A. thaliana encompassing all ORFs but discarding splice variants. Simply paste all Arabidopsis ORFs into the text field in the BiNGO Settings Panel, and select '---' in the test and correction fields. Name your 'cluster' e.g. A_thaliana_orfs. Select No visualization, All categories, GO_Full and Arabidopsis thaliana and press the start button. A custom annotation file called A_thaliana_orfs.anno is made.
How to make your own ontology files
In custom ontology files, the ontology categories are linked in a hierarchical structure as follows:
(curator=GO)(type=process)
0003673 = Gene_Ontology
0046087 = cytidine metabolism [isa: 0046131 ]
0046088 = cytidine biosynthesis [isa: 0046132 0046087 ]
0046089 = cytosine biosynthesis [isa: 0019856 0019858 ]
0042882 = L-arabinose transport [isa: 0015751 ]
0042883 = L-cysteine transport [isa: 0015807 ]
0042884 = microcin transport [isa: 0042891 0015833 ]
0045453 = bone resorption [partof: 0046849 ]
0042886 = amide transport [isa: 0006810 ]
0045452 = cuticle tanning [isa: 0040006 0008365 ] [partof: 0007562 ]
...
Again, the first line is obligatory. All subsequent lines contain an ontology category identifier linked to its description. If the category has parents in the hierarchy, these should be listed after [isa: , separated by space. [partof: is another type of hierarchical relationship defined in GO, e.g. 'mitotic cell cycle' isa kind of 'cell cycle', while the 'G2 phase' is partof the 'cell cycle'. Actually, BiNGO treats both types of relationship the same way, so it doesn't matter which one you use for ontologies that don't know these relationships, as long as you specify the [partof: part AFTER the [isa: part.
Copyright (c) 2005-2016 VIB


