|
|
|
|
|
|
||
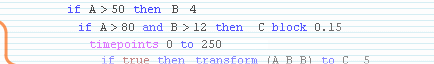
|
| Home |
| Tutorial | |
| Manual | |
| FAQ | |
| Thanks | |
| Download |
|
SIM-plex. Simulating genetic
networks made easy: network construction with simple building blocks |
||
| Welcome to
the homepage of the simulator that lets you define a genetic network
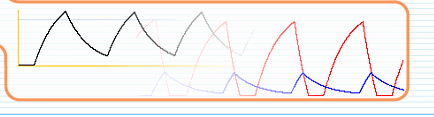
while avoiding complex biochemical differential equations! Genetic regulatory interactions activate a gene in a way that is often described by a sigmoid function. Already in 1973, Glass and Kauffman showed(1) that a network with this type of interactions can be replaced by a stepwise equivalent. In this 'substitute' network, the activations are approximated to happen switch-like or step-wise, while the outcome of the network behaviour will be the same as the original, continuous one. This allows us to avoid the use of biochemical
balance equations and to use simple stepwise functions instead, as a
first approximation. To get started with some examples, please read the tutorial section.
(1) Glass L and Kauffman SA. The logical analysis of continuous, nonlinear biochemical control networks. Journal of Theoretical Biology 39, 103-129 (1973).
|
